Abstract
The genomes of Bhanja virus (BHAV) and Palma virus (PALV) two tick-borne viruses hitherto grouped into the Bhanja virus antigenic complex of the Bunyaviridae were determined by pyrosequencing. Phylogenetic analysis groups all three segments of BHAV and PALV into a distinct clade of tick-borne phleboviruses together with the newly described severe fever with thrombocytopenia syndrome virus and Uukuniemi virus. The terminal signature sequences which are signatures for taxonomic grouping and important for virus replication and RNA transcription show marked differences in the L- and S-segments.
Similar content being viewed by others
Introduction
Bhanja virus (BHAV) was isolated in India in 1954 from a Haemaphysalis intermedia tick collected from a goat with paralytic symptoms. It has since been isolated in several countries in Africa and Europe from several tick species [13, 16, 26].
Serostudies detected BHAV antibodies in domestic cattle, sheep and goats, and man, in the African hedgehog (Atelerix albiventris) and the African ground squirrel (Xerus erythropus) [13]. More studies indicated the presence of BHAV in central Europe [15, 17, 24] around the Mediterranean basin, across the middle-eastern countries to India and in subsaharan Africa [4, 8, 22]. However, the strains north of the Sahara are serological distinct from the subsaharan strains [14]. BHAV has been described as pathogenic for young ruminants and as causing febrile and encephalitic disease in man. Animal models have confirmed it as a neurotropic virus [1, 12].
In 1992, a new bunyavirus called Palma virus (PALV) was isolated from a Haemaphysalis punctata adult tick pool collected from healthy cattle on a farm in Portugal. Morphological studies by electron microscopy and analysis of antigenic relationships by hemagglutination inhibition assays, immunofluorescence assays and plaque-reduction neutralisation assays in relation to BHAV, Kismaoa virus (KISV) and Forecariah virus (FOV) characterised this new virus as a member of the unassigned Bhanja antigenic group [10].
In 2009, a severe fever with thrombocytopenia syndrome was described in China and SFTSV was subsequently isolated from blood of patients that had been bitten by ticks [3, 27]. Complete genome analysis revealed it to be a Phlebovirus, which was then also detected in the tick Haemaphysalis longicornis [3, 27]. Person-to-Person transmission was described to people caring for infected patients [2, 11, 19].
We here describe the characterisation of the complete genome sequences of PALV and BHAV viruses and phylogenetic analysis of the determined sequences, which group into a clade with SFTSV and Uukuniemi virus (UUKV) identified in 1959 [23] and ever since used as a model for the analysis of the molecular biology of bunyaviruses.
Materials and methods
Growth and purification of Bhanja virus and Palma virus
Both virus isolates were from the Center for Vectors and Infectious Diseases Research, National Institute of Health, Portugal. BHAV M3811 was passaged three times in Vero E6 cells maintained in DMEM medium and supplemented with 2 % foetal bovine serum, 2 mM glutamine, 10 mM penicillin and 10 mM streptomycin in 175-cm2 tissue culture flasks. PALV M3443 was passaged three times in Vero B4 cells maintained in MEM medium supplemented with 2 % foetal bovine serum, 2 mM glutamine, 10 mM penicillin and 10 mM streptomycin in 175-cm2 tissue culture flasks. The medium of infected cells was collected when the cells showed 90–100 % CPE (5 dpi). Culture fluids were clarified by centrifugation at 700×g (2,000 rpm) for 10 min followed by an centrifugation at 2,800×g (4,000 rpm) for 5 min. Next the supernatants were filtered through a 0.2-μm sterile philtre. Twenty millilitre supernatant were mixed with 1.48 ml 5 M NaCl and 10.8 ml 30 % PEG8000 in NTE (10 mM Tris, pH 6.5; 1 mM EDTA; 100 mM NaCl) and incubated on a shaker for 30 min at 4 °C and subsequently centrifuged at 6,000 rpm for 60 min and 4 °C.
Pyrosequencing and bioinformatic analysis were performed as described [9], with the only exception that in addition to an 3′-FLAC adapter a 5′-RACE RNA adapter was ligated to the viral RNA after removal of two phosphate groups via RNA 5′-polyphosphatase treatment (FirstChoice RLM-RACE Kit Ambion). Dendrograms were made using Dendroscope [18].
Results
The complete genomes of BHAV and PALV were sequenced in one lane of a 4-lane picotiter plate in a pool of seven MID-tagged virus libraries in one pyrosequencing run. Each genome consists of three segments of negative-stranded ssRNA with a genome size of 11,554 and 11,549 bp, respectively (accession numbers: BHAV: JQ956376, JQ956377, JQ956378, PALV JQ956379, JQ956380, JQ956381). The coverage of the genome segments was about 41-fold (66 % of 2,641 reads) and 520-fold (47 % of 56,310 reads), respectively.
The length of the three BHAV and PALV genome segments, the calculated theoretical molecular mass of the encoded viral proteins and the nucleotide identities of BHAV vs PALV are detailed in Table 1.
For BHAV and PALV, the 5′ and 3′ NCR sequences are different for each genome segment. While the M-segment carries the typical phlebovirus signature, the L-segment has varying 5′- and 3′-signatures and the S-segment shows a variant 3′-signature sequence (see Table 2).
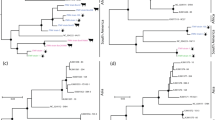
Phylogenetic analysis in comparison to 134, 132 and 196 full length L-, M-, and S-phlebovirus sequences clearly grouped BHAV and PALV with SFTSV and UUKV supported by high bootstrap values (see Fig. 1).
a–c Phylogenetic trees of 142 L-, 134 M-, and 169 S-segments. Analysis of nucleotide sequences was performed using ClustalW and a 1000-fold bootstrap approach rooted to the sequence of the Orthobunyavirus Bunyamwera virus. Bootstrap values are given in percent. Genus, Phlebovirus species, Phlebovirus tentative species (italic grey), virus (number of sequences included). To simplify the dendrogram for presentation subtrees were collapsed. Full dendrograms can be viewed in the supporting material. Tick-borne phleboviruses on red branches. c One consensus sequence representing 85 RVFV sequences was used for the simplified S-dendrogram. The unassigned viruses in c are published phlebovirus sequences from South America that have not yet been assigned to a genus: Phlebovirus BeAn578142, BeAn 416992, BeAn356637, BeAn24262, VP-161A, BeAn 371637 (see full dendrogram in supporting material) (Color figure online)
Discussion
BHAV has so far had the status of an unassigned candidate member of the Bunyaviridae virus family and is currently not recorded as a member or tentative member of the genus Phlebovirus. Phylogenetic analysis of the newly determined L- and M-segment sequences of BHAV and PALV, clades them into a single subtree together with the new tick-borne phlebovirus SFTSV from China and UUKV within the genus Phlebovirus of the Bunyaviridae.
The subtree for the S-segment sequences of these tick-borne viruses is not as clear cut as it also contains the recently determined sequences of 13 mosquito borne viruses from South America (Candiru virus) [21] which in contrast group into distinct subtrees in the L- and M-segment dendrograms.
The difference among the tick-borne viruses grouping together is quite high suggesting a long evolutionary history in the slow evolutionary process described for viruses in ticks [20, 28]. It is of note that UUKV and SFSTV are more closely related to each other than to either BHAV or PALV (Fig. 1a–c, depth of branches, Table 3).
The BHAV antigenic group into which PALV was placed by serology was originally difficult to assign and this may stem from the deviant nature of the surface glycoprotein genes of BHAV and PALV. The amino acid sequence of the respective glycoprotein precursors share 91.1 % identity to each other but only 20.9 and 21.1 % to UUKV which was clearly identified as a Phlebovirus [25]. However, all three dendrograms clearly group the new BHAV and PALV sequences inside the genus Phlebovirus and therefore allow to suggest that both BHAV and PALV should be placed into the genus Phlebovirus.
Inverted signature sequences in the terminal non coding-region which allow panhandle formation of the ssRNA segments have been described for each genus in the Bunyaviridae family. A differing signature for isolates of SFTSV recently described in China is the first case challenging the observation that there are conserved signature sequences for each genus. This observation can now be extended by the differences observed in the terminal L- and S-segment signatures of BHAV and PALV. Whether this has any impact on the efficiency of replication [5], or RNA synthesis [6, 7] during the replication cycle of theses viruses remains to be seen. These particularities are another indication of the divergent nature of BHAV and PALV within the genus Phlebovirus.
References
M. Balducci, P. Verani, M.C. Lopes, F. Nardi, Experimental pathogenicity of Bhanja virus for white mice and Macaca mulatta monkeys. Acta Virol. 14(3), 237–243 (1970)
C.J. Bao, X.L. Guo, X. Qi, J.L. Hu, M.H. Zhou, J.K. Varma, L.B. Cui, H.T. Yang, Y.J. Jiao, J.D. Klena, L.X. Li, W.Y. Tao, X. Li, Y. Chen, Z. Zhu, K. Xu, A.H. Shen, T. Wu, H.Y. Peng, Z.F. Li, J. Shan, Z.Y. Shi, H. Wang, A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin. Infect. Dis. 53(12), 1208–1214 (2011)
C.J. Bao, X. Qi, H. Wang, A novel bunyavirus in China. N. Engl. J. Med. 365(9), 862–863 (2011). (author reply 864–865)
V. Bardos, W. Sixl, C.L. Wisidagama, J. Halouzka, D. Stunzner, Z. Hubalek, H. Withalm, Prevalence of arbovirus antibodies in sera of animals in Sri Lanka. Bull. World Health Organ. 61(6), 987–990 (1983)
J.N. Barr, R.M. Elliott, E.F. Dunn, G.W. Wertz, Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 311(2), 326–338 (2003)
J.N. Barr, G.W. Wertz, Bunyamwera bunyavirus RNA synthesis requires cooperation of 3′- and 5′-terminal sequences. J. Virol. 78(3), 1129–1138 (2004)
J.N. Barr, G.W. Wertz, Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 79(6), 3586–3594 (2005)
M.A. Darwish, H. Hoogstraal, T.J. Roberts, R. Ghazi, T. Amer, A sero-epidemiological survey for Bunyaviridae and certain other arboviruses in Pakistan. Trans. R. Soc. Trop. Med. Hyg. 77(4), 446–450 (1983)
M. Dilcher, L. Hasib, M. Lechner, N. Wieseke, M. Middendorf, M. Marz, A. Koch, M. Spiegel, G. Dobler, F.T. Hufert, M. Weidmann, Genetic characterization of Tribec virus and Kemerovo virus, two tick-transmitted human-pathogenic Orbiviruses. Virology 423(1), 68–76 (2012)
A.R. Filipe, M.J. Alves, N. Karabatsos, A.P. de Matos, M.S. Nuncio, F. Bacellar, Palma virus, a new bunyaviridae isolated from ticks in Portugal. Intervirology 37(6), 348–351 (1994)
Z. Gai, M. Liang, Y. Zhang, S. Zhang, C. Jin, S.W. Wang, L. Sun, N. Zhou, Q. Zhang, Y. Sun, S.J. Ding, C. Li, W. Gu, F. Zhang, Y. Wang, P. Bian, X. Li, Z. Wang, X. Song, X. Wang, A. Xu, Z. Bi, S. Chen, D. Li, Person-to-person transmission of severe fever with thrombocytopenia syndrome bunyavirus through blood contact. Clin. Infect. Dis. 54(2), 249–252 (2012)
Z. Hubalek, Experimental pathogenicity of Bhanja virus. Zentralbl Bakteriol Mikrobiol Hyg A 266(1–2), 284–291 (1987)
Z. Hubalek, Geographic distribution of Bhanja virus. Folia Parasitol. (Praha) 34(1), 77–86 (1987)
Z. Hubalek, J. Halouzka, Numerical comparative serology of the Bhanja antigenic group (Bunyaviridae). Arch. Virol. 84(3–4), 175–180 (1985)
Z. Hubalek, Z. Juricova, A serological survey for Bhanja virus in Czechoslovakia. Zentralbl Bakteriol Mikrobiol Hyg A 258(4), 540–543 (1984)
Z. Hubalek, T. Mittermayer, J. Halouzka, V. Cerny, Isolation of “exotic” Bhanja virus (Bunyaviridae) from ticks in the temperate zone. Arch. Virol. 101(3–4), 191–197 (1988)
Z. Hubalek, J. Mitterpak, J. Prokopic, Z. Juricova, J. Kilik, A serological survey for Bhanja and tick-borne encephalitis viruses in sheep of eastern Slovakia. Folia Parasitol. (Praha) 32(3), 279–283 (1985)
D.H. Huson, D.C. Richter, C. Rausch, T. Dezulian, M. Franz, R. Rupp, Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 480 (2007)
Y. Liu, Q. Li, W. Hu, J. Wu, Y. Wang, L. Mei, D.H. Walker, J. Ren, Y. Wang, X.J. Yu, Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 12(2), 156–160 (2012)
M.S. Marin, P.M. Zanotto, T.S. Gritsun, E.A. Gould, Phylogeny of TYU, SRE, and CFA virus: different evolutionary rates in the genus Flavivirus. Virology 206(2), 1133–1139 (1995)
G. Palacios, R. Tesh, A.T. Da Rosa, N. Savji, W. Sze, K. Jain, R. Serge, H. Guzman, C. Guevara, M.R. Nunes, J.P. Nunes-Neto, T. Kochel, S. Hutchison, P.F. Vasconcelos, W.I. Lipkin, Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J. Virol. 85(8), 3811–3820 (2011)
V. Punda, D. Ropac, J. Vesenjak-Hirjan, Incidence of hemagglutination-inhibiting antibodies for Bhanja virus in humans along the north-west border of Yugoslavia. Zentralbl Bakteriol Mikrobiol Hyg A 265(1–2), 227–234 (1987)
P. Saikku, C.H. von Bonsdorff, Electron microscopy of the Uukuniemi virus, an ungrouped arbovirus. Virology 34(4), 804–806 (1968)
S. Sikutova, S. Hornok, Z. Hubalek, I. Dolezalkova, Z. Juricova, I. Rudolf, Serological survey of domestic animals for tick-borne encephalitis and Bhanja viruses in northeastern Hungary. Vet. Microbiol. 135(3–4), 267–271 (2009)
J.F. Simons, U. Hellman, R.F. Pettersson, Uukuniemi virus S RNA segment: ambisense coding strategy, packaging of complementary strands into virions, and homology to members of the genus Phlebovirus. J. Virol. 64(1), 247–255 (1990)
A. Ungureanu, V. Popovici, F. Catanas, I. Ionita, A. Tutoveanu, M. Safta, Isolation of Bhanja virus in Romania. Arch. Roum. Pathol. Exp. Microbiol. 49(2), 139–145 (1990)
X.J. Yu, M.F. Liang, S.Y. Zhang, Y. Liu, J.D. Li, Y.L. Sun, L. Zhang, Q.F. Zhang, V.L. Popov, C. Li, J. Qu, Q. Li, Y.P. Zhang, R. Hai, W. Wu, Q. Wang, F.X. Zhan, X.J. Wang, B. Kan, S.W. Wang, K.L. Wan, H.Q. Jing, J.X. Lu, W.W. Yin, H. Zhou, X.H. Guan, J.F. Liu, Z.Q. Bi, G.H. Liu, J. Ren, H. Wang, Z. Zhao, J.D. Song, J.R. He, T. Wan, J.S. Zhang, X.P. Fu, L.N. Sun, X.P. Dong, Z.J. Feng, W.Z. Yang, T. Hong, Y. Zhang, D.H. Walker, Y. Wang, D.X. Li, Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364(16), 1523–1532 (2011)
P.M. Zanotto, G.F. Gao, T. Gritsun, M.S. Marin, W.R. Jiang, K. Venugopal, H.W. Reid, E.A. Gould, An arbovirus cline across the northern hemisphere. Virology 210(1), 152–159 (1995)
Acknowledgments
This study was supported by the Federal Ministry of Education and Research (BMBF), Grant Number 01KI0710, ‘Research on Zoonotic Infectious Diseases’ Programme, ‘Emerging arthropode-borne viral infections in Germany: Pathogenesis, diagnostics and surveillance’ and the BMBF funded research programme ‘Potential release-oriented biothreat emergency diagnostics (P.R.O.B.E)’ for civil security of the German Federal Government as part of the high-tech strategy for Germany.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Dilcher, M., Alves, M.J., Finkeisen, D. et al. Genetic characterization of Bhanja virus and Palma virus, two tick-borne phleboviruses. Virus Genes 45, 311–315 (2012). https://doi.org/10.1007/s11262-012-0785-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-012-0785-y