Abstract
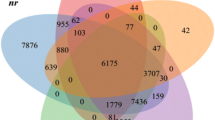
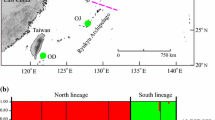
Lindera glauca (Lauraceae) is an economically important East Asian forest tree characterized by a dioecy in China and apomixis in Japan. However, patterns of population genetic diversity and structure of this species remain unknown for this species due to a lack of efficient molecular markers. In this study, we employed Illumina sequencing to analyze the transcriptomes of the female and male flower buds of L. glauca. We retrieved 59,753 and 75,075 unigenes for the female and male buds, respectively. Based on sequence similarity, 44,379 (74.27 %) unigenes for the female and 45,414 (60.49 %) unigenes for the male were matched to public databases. We identified 11,127 putative differentially expressed genes between the female and male buds and 20,048 expressed sequence tag-simple sequence repeats (EST-SSRs). From 3147 primer pairs designed successfully, 120 were selected for validation of polymorphism, and 13 could reliably amplify polymorphic bands and exhibited moderate levels of genetic diversity (e.g., N A = 4.42; H E = 0.56) when surveyed across 96 individuals of altogether six L. glauca populations from China and Japan. One of the three population genetic clusters identified in China was fixed in Japan, suggesting a historical population bottleneck following island immigration. The present study has generated a wealth of transcriptome data for future functional genomic research focused on the variable reproductive system of L. glauca (dioecy, apomixis) as well as EST-SSR markers for population genetics studies and its intriguing evolutionary shift from dioecy to apomixis in the wake of island colonization.


Similar content being viewed by others
References
Wang YL, Gao XM, Yu XP, Cheng SL, Kong LH (1994) Study on the resource and its utilizations of Lindera glauca in China. Henan Sci 12:331–334
Niu J, Chen YL, An JY, Hou XY, Cai J, Wang J, Zhang ZX, Lin SZ (2015) Integrated transcriptome sequencing and dynamic analysis reveal carbon source partitioning between terpenoid and oil accumulation in developing Lindera glauca fruits. Sci Rep 5:15017
Qi J, Xiong B, Ju YX, Hao QZ, Zhang ZX (2015) Study on fruit growth regularity and lipid accumulation of Lindera glauca. Chin Agric Sci Bull 31:29–33
Dupont YL (2002) Evolution of apomixis as a strategy of colonization in the dioecious species Lindera glauca (Lauraceae). Popul Ecol 44:0293–0297
Pannell JR, Dorken ME (2006) Colonisation as a common denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landsc Ecol 21:837–848
Rodríguez-Sánchez F, Guzmán B, Valido A, Vargas P, Arroyo J (2009) Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. J Biogeogr 36:1270–1281
Niu J, Hou XY, Fang CL, An JY, Ha DL, Qiu L, Ju YX, Zhao HY, Du WZ, Qi J, Zhang ZX, Liu GN, Lin SZ (2015) Transcriptome analysis of distinct Lindera glauca tissues revealed the differences in the unigenes related to terpenoid biosynthesis. Gene 559:22–30
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Guo R, Mao YR, Cai JR, Wang JY, Wu J, Qiu YX (2014) Characterization and cross-species transferability of EST-SSR markers developed from the transcriptome of Dysosma versipellis (Berberidaceae) and their application to population genetic studies. Mol Breed 34:1733–1746
Chen LY, Cao YN, Yuan N, Nakamura K, Wang GM, Qiu YX (2015) Characterization of transcriptome and development of novel EST-SSR makers based on next-generation sequencing technology in Neolitsea sericea (Lauraceae) endemic to East Asian land-bridge islands. Mol Breed 35:1–15
Ai B, Gao Y, Zhang X, Tao J, Kang M, Huang H (2015) Comparative transcriptome resources of eleven Primulina species, a group of ‘stone plants’ from a biodiversity hot spot. Mol Ecol Resour 15:619–632
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Zhou XJ, Wang YY, Xu YN, Yan RS, Zhao P, Liu WZ (2015) De novo characterization of flower bud transcriptomes and the development of EST-SSR markers for the endangered tree Tapiscia sinensis. Int J Mol Sci 16:12855–12870
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B (2003) TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19:651–652
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Zhang J, Feng JJ, Lu J, Yang YZ, Zhang X, Wan DS, Liu JQ (2014) Transcriptome differences between two sister desert poplar species under salt stress. BMC Genomics 15:1
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices Version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Takezaki N, Nei M, Tamura K (2010) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27:747–752
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Jiang X, Wu Y, Xiao F, Xiong Z, Xu H (2014) Transcriptome analysis for leaves of five chemical types in Cinnamomum camphora. Hereditas 36:58–68
Mao YR, Zhang YH, Xu C, Qiu YX (2015) Comparative transcriptome resources of two Dysosma species (Berberidaceae) and molecular evolution of the CYP719A gene in Podophylloideae. Mol Ecol Resour 16:228–241
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Stoeckel S, Grange J, Fernandez-Manjarres F, Bilger I, Frascaria-Lacoste N, Mariette S (2006) Heterozygote excess in a self-incompatible and partially clonal forest tree species—Prunus avium L. Mol Ecol 15:2109–2118
Barrett SCH (1996) The reproductive biology and genetics of island plants. Philos Trans R Soc Lond B 351:725–733
Ronsheim ML, Bever JD (2000) Genetic variation and evolutionary trade-offs for sexual and asexual reproductive modes in Allium vineale (Liliaceae). Am J Bot 87:1769–1777
Obbard DJ, Harris SA, Pannell JR (2006) Sexual systems and population genetic structure in an annual plant: testing the metapopulation model. Am Nat 167:354–366
Yuan N, Sun Y, Comes HP, Fu CX, Qiu YX (2014) Understanding population structure and historical demography in a conservation context: population genetics of the endangered Kirengeshoma palmata (Hydrangeaceae). Am J Bot 101:521–529
Barrett SCH (2002) The evolution of plant sexual diversity. Nat Rev Gen 3:274–284
Acknowledgments
The research was supported by the National Science Foundation of China (Grant No. 3151101152) and the International Cooperation and Exchange of the National Natural Science Foundation of China (Grant Nos. 31511140095, 31561143015). The authors are grateful to Koh Nakamura (Hokkaido University, Japan) for collecting Japanese material of L. glauca, Hans-Peter Comes and two anonymous reviewers for very insightful comments on an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, S., Ding, Y., Yap, Z. et al. De novo assembly and characterization of the floral transcriptome of an economically important tree species, Lindera glauca (Lauraceae), including the development of EST-SSR markers for population genetics. Mol Biol Rep 43, 1243–1250 (2016). https://doi.org/10.1007/s11033-016-4056-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4056-1