Abstract
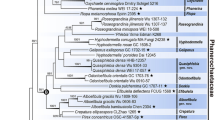
The aims of this study are to assess the utility of the internal transcribed spacer (ITS) region, and partial translation elongation factor (EF1α) and RNA polymerase II (RPB2) genes, for differentiation of Bailinggu, P. eryngii, and P. nebrodensis; to reconstruct phylogenetic relationships between the three species; and to confirm the taxonomic status of Bailinggu based on ribosomal and protein-coding genes. Pairwise genetic distances between Bailinggu, P. eryngii, and related Pleurotus strains were calculated by using the p-distance model, and molecular phylogeny of these isolates was estimated based on ITS, RPB2, and EF1α using maximum parsimony and Bayesian methods. Differences in ITS, RPB2, and EF1α sequences show that Bailinggu, P. eryngii, and P. nebrodensis are distinct at the species level. Phylogenetic analyses reveal that P. eryngii is closer to P. nebrodensis than to Bailinggu. Sequence analyses of ribosomal and protein-coding genes confirm that P. eryngii var. tuoliensis is identical to Bailinggu. P. eryngii var. tuoliensis should be raised to species level or a new name should be introduced for Bailinggu after a thorough investigation into Pleurotus isolates from Ferula in Xinjiang Province. This study helps to resolve uncertainty regarding Bailinggu, P. eryngii and P. nebrodensis, improving the resource management of these strains. ITS, EF1α, and RPB2 sequences can be used to distinguish Bailinggu, P. eryngii and P. nebrodensis as three different species, and P. eryngii var. tuoliensis should be the scientific name for Bailinggu at present.



Similar content being viewed by others
References
Mao XL (2000) Agaricales. In: Mao XL (ed) The macrofungi in China. Henan Science and Technology Press, Zhengzhou, pp 64–66
Mao X (2005) Promoting a new development for precious mushroom Pleurotus nebrodensis (in Chinese). China (Guang Shui) Symposium on Standardization Production for Edible Mushroom & Products Fair for Rare Mushroom (Pleurotus nebrodensis), Hubei, China, January 17–18, pp 25–27 (in Chinese)
Zhang J, Huang C, Li C (2005) The cultivars of P. nebrodensis in China. In: Tan Q, Zhang J, Chen M, Cao H, Buswell JA (eds) Mushroom biology and mushroom products, vol 12. Shanghai Xinhua Printing Co., Ltd, Acta Edulis Fungi, Shanghai, pp 350–353
Huang N (1998) Colored illustrations of macrofungi (mushrooms) of China. China Agricultural Press, Beijing, p 96 (in Chinese)
Kawai G, Babasaki K, Neda H (2008) Taxonomic position of a Chinese Pleurotus ‘‘Bailinggu’’: it belongs to Pleurotus eryngii (DC.: Fr.) Quél. and evolved independently in China. Mycoscience 49:75–87
Zhao M, Huang C, Chen Q, Wu X, Qu J, Zhang J (2013) Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 8:e83253
Jia SM, Qin M (2006) Domestication and cultivation of Pleurotus nebrodensis in China. Edible Fungi China 25:3–7 (in Chinese)
Xu ML (2010) Sexual distant-crossbreeding between P. eryngii and P. nebrodensis. Master’s Thesis, Fujian Agriculture and Forestry University
Li GX, Shao SG, Li YJ (2004) Effects of different hormones on growth and yield of Pleurotus eryngii var. nebrodensis. Edible Fungi of China 23:37–38
Xu JY (2010) The preliminary study on the structure of A mating type loci in Pleurotus eryngii var. nebrodensis and Pleurotus eryngii var. ferulae. Master’s Thesis, Huazhong Agricultural University
Mou C, Cao Y, Ma J (1987) A new variety of Pleurotus eryngii and its cultural characters. Acta Mycol Sin 6:153–156 (in Chinese)
Zhang JX, Huang CY, Ng TB, Wang HX (2006) Genetic polymorphism of ferula mushroomgrown on Ferula sinkiangensis. Appl Microbiol Biotechnol 71:304–309
Bao D, Kinugasa S, Kitamoto Y (2004) The biological species of oyster mushrooms (Pleurotus spp.) from Asia based on mating compatibility tests. J Wood Sci 50:162–168
Bao D, Ishihara H, Mori N, Kitamoto Y (2004) Phylogenetic analysis of oyster mushrooms (Pleurotus spp.) based on restriction fragment length polymorphisms of the 5′ portion of 26S rDNA. J Wood Sci 50:169–176
Ro HS, Kim SS, Ryu JS, Jeon CO, Lee TS, Lee HS (2007) Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol Res 111:710–715
Rodriguez Estrada AE (2008) Molecular phylogeny and increases of yield and the antioxidants selenium and ergothioneine in basidiomata of Pleurotus eryngii. Ph.D. Dissertation, The Pennsylvania State University
Avin FA, Bhassu S, Tan YS, Shahbazi P, Vikineswary S (2014) Molecular divergence and species delimitation of the cultivated oyster mushrooms: integration of IGS1 and ITS. Sci World J. doi:10.1155/2014/793414
Froslev TG, Matheny PB, Hibbett DS (2005) Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): a comparison of RPB1, RPB2, and ITS phylogenies. Mol Phylogenet Evol 37:602–618
Rodriguez Estrada AE, del Mar Jimenez-Gasco M, Royse DJ (2010) Pleurotus eryngii species complex: sequence analysis and phylogeny based on partial EF1α and RPB2 genes. Fungal Biol 114:421–428
Schoch CL, Seifert K, Huhndorf S et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 109:241–6246
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego
Liu YJ, Whelen S, Hall D (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Matheny PB (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol 35:1–20
Marongiu P, Maddau L, Frisullo S, Marras F (2005) A multigene approach for the taxonomic determination of Pleurotus eryngii isolates. In: Tan Q, Zhang J, Chen M, Cao H, Buswell JA (eds) Mushroom Biology and Mushroom Products, vol 12. Shanghai Xinhua Printing Co., Ltd, Acta Edulis Fungi, Shanghai, pp 89–91
Wendland J, Kothe E (1997) Isolation of tef1 encoding translation elongation factor EF-1α from the homobasidiomycete Schizophyllum commune. Mycol Res 101:798–802
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10. Sinauer, Sunderland
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Ravash R, Shiran B, Alavi A-A, Bayat F, Rajaee S, Zervakis GI (2010) Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Prog 9:181–194
Zervakis G, Venturella G, Papadopoulou K (2001) Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species complex as determined by RAPD analysis, isozyme profiles and ecomorphological characters. Microbiology 147:3183–3194
Zervakis G, Balis C (1996) A pluralistic approach on the study of Pleurotus species, with emphasis on compatibility and physiology of the European morphotaxa. Mycol Res 100:717–731
Venturella G (2000) Typification of Pleurotus nebrodensis. Mycotaxon 75:229–231
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Acknowledgments
Dr. Egon Horak is acknowledged for valuable suggestions to improve the manuscript. The research was financed by the Applied Basic Research, Science and Technology Department of Sichuan Province (Project No. 2013JY0114), National Public Welfare (Agriculture) Science and Technology Project (201503137), Sichuan Provincial Infrastructure of Microbial Resources (15010302) and Sichuan Provincial Innovation Ability Promotion Engineering (Project No. 2014LWJJ-005).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
He, XL., Wu, B., Li, Q. et al. Phylogenetic relationship of two popular edible Pleurotus in China, Bailinggu (P. eryngii var. tuoliensis) and Xingbaogu (P. eryngii), determined by ITS, RPB2 and EF1α sequences. Mol Biol Rep 43, 573–582 (2016). https://doi.org/10.1007/s11033-016-3982-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3982-2