Abstract
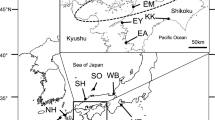
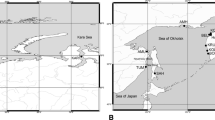
Marine pelagic fishes are usually characterized by subtle but complex patterns of genetic differentiation, which are influenced by both historical process and contemporary gene flow. Genetic population differentiation of chub mackerel, Scomber japonicus, was examined across most of its range in the Northwestern Pacific by screening variation of eight microsatellite loci. Our genetic analysis detected a weak but significant genetic structure of chub mackerel, which was characterized by areas of gene flow and isolation by distance. Consistent with previous estimates of stock structure, we found genetic discontinuity between Japan and China samples. Local-scale pattern of genetic differentiation was observed between samples from the Bohai Sea and North Yellow Sea and those from the East China Sea, which we ascribed to differences in spawning time and migratory behavior. Furthermore, the observed homogeneity among collections of chub mackerel from the East and South China Seas could be the result of an interaction between biological characteristics and marine currents. The present study underlies the importance of understanding the biological significance of genetic differentiation to establish management strategies for exploited fish populations.




Similar content being viewed by others
References
Palumbi SR (1994) Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst 25:547–572
Arnason E, Pálsson S (1996) Mitochondrial cytochrome b DNA sequence variation of Atlantic cod Gadhus morhua, from Norway. Mol Ecol 5:715–724
Vis ML, Carr SM, Bowering WR, Davidson WS (1997) Greenland halibut (Reinhartius hippoglossides) in the North Atlantic are genetically homogeneous. Can J Fish Aquat Sci 54:1813–1821
Smedbol RK, McPerson A, Hansen MM, Kenchington E (2002) Myths and moderation in marine ‘metapopulations’? Fish Fish 3:20–35
Nielsen EE, Nielsen PH, Meldrup D, Hansen MM (2004) Genetic population structure of turbot (Scophthalmus maximus L.) supports the presence of multiple hybrid zones for marine fishes in the transition zone between the Baltic Sea and the North Sea. Mol Ecol 13:585–595
Bekkevold D, André C, Dahlgren TG, Clausen LA, Torstensen E, Mosegaard H, Carvalho GR, Christensen TB, Norlinder E, Ruzzante DE (2005) Environmental correlates of population differentiation in Atlantic herring. Evolution 59:2656–2668
Was A, Gosling E, McCrann K, Mork J (2008) Evidence for population structuring of blue whiting (Micromesistius poutassou) in the Northeast Atlantic. ICES J Mar Sci 65:216–225
Cunningham KM, Canino MF, Spies IB, Hauser L (2009) Genetic isolation by distance and localized fjord population structure in Pacific cod (Gadus macrocephalus): limited effective dispersal in the northeastern Pacific Ocean. Can J Fish Aquat Sci 66:153–166
André C, Larsson LC, Laikre L, Bekkevold D, Brigham J, Carvalho GR, Dahlgren TG, Hutchinson WF, Mariani S, Mudde K, Ruzzante DE, Ryman N (2011) Detecting population structure in a high gene-flow species, Atlantic herring (Clupea harengus): direct, simultaneous evaluation of neutral vs putatively selected loci. Heredity 106:270–280
Viñas J, Alvarado Bremer J, Pla C (2004) Phylogeography of the Atlantic bonito (Sarda sarda) in the northern Mediterranean: the combined effects of historical vicariance, population expansion, secondary invasion, and isolation by distance. Mol Phylogenet Evol 33:32–42
Liu JX, Tatarenkov A, Beacham TD, Gorbachev V, Wildes S, Avise JC (2011) Effects of Pleistocene climatic fluctuations on the phylogeographic and demographic histories of Pacific herring (Clupea pallasii). Mol Ecol 20:3879–3893
Magoulas A, Castilho R, Caetano S, Marcato S, Patarnello T (2006) Mitochondrial DNA reveals a mosaic pattern of phylogeographical structure in Atlantic and Mediterranean populations of anchovy (Engraulis encrasicolus). Mol Phylogenet Evol 39:734–746
Jørgensen HB, Hansen MM, Bekkevold D, Ruzzante DE, Loeschcke V (2005) Marine landscapes and population genetic structure of herring (Clupea harengus L.) in the Baltic Sea. Mol Ecol 14:3219–3234
Han Z, Yanagimoto T, Zhang Y, Gao T (2012) Phylogeography study of Ammodytes personatus in Northwestern Pacific: pleistocene isolation, temperature and current conducted secondary contact. PLoS ONE 7(5):e37425. doi:10.1371/journal.pone.0037425
Zardoya R, Castilho R, Grande C, Favre-Krey L, Caetano S, Marcato S, Krey G, Patarnello T (2004) Differential population structuring of two closely related fish species, the mackerel (Scomber scombrus) and the chub mackerel (Scomber japonicus), in the Mediterranean Sea. Mol Ecol 13:1785–1798
Nesbø CL, Rueness EK, Iversen SA, Skagen DW, Jakobsen KS (2000) Phylogeography and population history of Atlantic mackerel (Scomber scombrus L.): a genealogical approach reveals genetic structuring among eastern Atlantic stocks. Proc R Soc Lond B 267:281–292
Natoli A, Birkun A, Aguilar A, Lopez A, Hoelzel AR (2005) Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc R Soc Lond B 272:1217–1226
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450
Avise JC (2000) Phylogeography: The history and formation of species. Harvard University Press, Cambridge
Carvalho GR, Hauser L (1994) Molecular genetics and the stock concept in fisheries. Rev Fish Biol Fish 4:326–350
Ward RD, Grewe PM (1994) Appraisal of molecular genetic techniques in fisheries. Rev Fish Biol Fish 4:300–325
Hutchings JA (2000) Collapse and recovery of marine fishes. Nature 406:882–885
Liu M, Lin LS, Gao TX, Yanagimoto T, Sakurai Y, Grant WS (2012) What maintains the central North Pacific genetic discontinuity in Pacific herring? PLoS ONE 7:e50340. doi:10.1371/journal.pone.0050340
Scoles DR, Collette BB, Graves JE (1998) Global phylogeography of mackerels of the genus Scomber. Fish Bull 96:823–842
Infante C, Blanco E, Zuasti E, Crespo A, Manchado M (2007) Phylogenetic differentiation between Atlantic Scomber colias and Pacific Scomber japonicus based on nuclear DNA sequences. Genetica 130:1–8
Catanese G, Manchado M, Infante C (2010) Evolutionary relatedness of mackerels of the genus Scomber based on complete mitochondrial genomes: Strong support to the recognition of Atlantic Scomber colias and Pacific Scomber japonicus as distinct species. Gene 452:35–43
Collette BB, Nauen CE (1983) Scombrids of the world: An annotated and illustrated catalogue of tunas, mackerels, bonitos and related species known to date. FAO Fish Synop 125:1–137
Li CS, Li YZ, Li GF, Chen WZ, Chen GB, Chen CH, Jin XS, Yu LF, Lin LY (2006) Pelagic fishes. In: Tang QS (ed) Marin biology resources and habitat environments in the EEZ of China. Science Press, Beijing, pp 599–608
Yukami R, Ohshimo S, Yoda M, Hiyama Y (2009) Estimation of the spawning grounds of chub mackerel Scomber japonicus and spotted mackerel Scomber australasicus in the East China Sea based on catch statistics and biometric data. Fish Sci 75:167–174
Tzeng TD, Haung HL, Wang D, Yeh SY (2007) Genetic diversity and population expansion of the common mackerel (Scomber japonicus) off Taiwan. J Fish Soc Taiwan 34:237–245
Shao F, Chen XJ (2008) Genetic variations of Scomber japonicus and S. australasicus based on random amplified polymorphic DNA in the Yellow Sea and the East China Sea. J Guangdong Ocean Univ 28:83–87 (in Chinese with English abstract)
Zhang LY, Su YQ, Wang HJ, Wang J (2011) Population genetic structure of Pneumatophorus japonicus in the Taiwan Strait. Acta Ecol Sin 31:7097–7103 (in Chinese with English abstract)
Zeng L, Cheng Q, Chen X (2012) Microsatellite analysis reveals the population structure and migration patterns of Scomber japonicus (Scombridae) with continuous distribution in the East and South China Seas. Biochem Syst Ecol 42:83–93
Ruzzante DE, Taggart CT, Cook D (1999) A review of the evidence for genetic structure of cod (Gadus morhua) populations in the NW Atlantic and population affinities of larval cod off Newfoundland and the Gulf of St Lawrence. Fish Res 43:79–97
Nielsen EE, Hansen MM, Schmidt C, Meldrup D, Grønkjaer P (2001) Population of origin of Atlantic cod. Nature 413:272
Carlsson J, McDowell JR, Diaz-Jaimes P, Carlsson JE, Bole SB, Gold JR, Graves J (2004) Microsatellite and mitochondrial DNA analyses of Atlantic bluefin tuna (Thunnus thynnus thynnus) population structure in the Mediterranean Sea. Mol Ecol 13:3345–3356
Matsui T (1967) Review of the mackerel genera Scomber and Rastrelliger with description of a new species of Rastrelliger. Copeia 1:71–83
Sambrook J, Russell DW (2001) Molecular cloning: A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Yagishita N, Kobayashi T (2008) Isolation and characterization of nine microsatellite loci from the chub mackerel, Scomber japonicus (Perciformes, Scombridae). Mol Ecol Resour 8:302–304
Tang CY, Tzeng CH, Chen CS, Chiu TS (2009) Microsatellite DNA markers for population genetic studies of blue mackerel (Scomber australasicus) and cross-specific amplification in S. japonicus. Mol Ecol Resour 9:824–827
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 19:680–683
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) POPGENE 32, Microsoft Windows-based freeware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Goudet J (2001) FSTAT V.2.9.3, a program to estimate and test gene diversities and fixation indices (Updated from Goudet 1995). University of Lausanne, Lausanne
Raymond M, Rousset F (1995) GENEPOP Version 1.2: population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analysing tables of statistical tests. Evolution 43:223–225
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 130:457–462
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, Université de Montpellier II, Montpellier
Pritchard JK, tephens MS, onnelly PD (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN, Version 2.0: a software for population genetic data analysis. University of Geneva, Geneva
Cárdenas L, Silva AX, Magoulas A, Cabezas J, Poulin E, Ojeda FP (2009) Genetic population structure in the Chilean jack mackerel, Trachurus murphyi (Nichols) across the South-eastern Pacific Ocean. Fish Res 100:109–115
Fauvelot C, Borsa P (2011) Patterns of genetic isolation in a widely distributed pelagic fish, the narrow-barred Spanish mackerel (Scomberomorus commerson). Biol J Linn Soc 104:886–902
Ross KG, Krieger MJ, Shoemaker DD, Vargo EL, Keller L (1997) Hierarchical analysis of genetic structure in native fire ant populations: results from three classes of molecular markers. Genetics 147:643–655
Estoup A, Angers B (1998) Microsatellites and minisatellites for molecular ecology: theoretical and empirical considerations. In: Carvalho GR (ed) Advances in molecular ecology. IOS Press, Amsterdam, pp 55–86
Templeton A, Routman E, Phillips C (1995) Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140:767–782
Templeton A (2001) Using phylogeographic analyses of gene trees to test species status and processes. Mol Ecol 10:779–791
McQuinn IH (1997) Metapopulations and the Atlantic herring. Rev Fish Biol Fish 7:297–329
Knutsen H, Jorde PE, André C, Stenseth NC (2003) Fine-scaled geographical population structuring in a highly mobile marine species: the Atlantic cod. Mol Ecol 12:385–394
Shiraishi T, Okamoto K, Yoneda M, Sakai T, Ohshimo S, Onoe S, Yamaguchi A, Matsuyama M (2008) Age validation, growth and annual reproductive cycle of chub mackerel Scomber japonicus off the waters of northern Kyushu and in the East China Sea. Fish Sci 74:947–954
Tzeng TD (2007) Population structure and historical demography of the spotted mackerel (Scomber australasicus) off Taiwan inferred from mitochondrial control region sequencing. Zool Stud 46:656–663
Zhan A, Hu J, Hu X, Zhou Z, Hui M, Wang S, Peng W, Wang M, Bao Z (2009) Fine-scale population genetic structure of Zhikong scallop (Chlamys farreri): do local marine currents drive geographical differentiation? Mar Biotechnol 11:223–235
Liu M, Lu ZC, Gao TX, Yanagimoto T, Sakurai Y (2010) Remarkably low mtDNA control-region diversity and shallow population structure in Pacific cod Gadus macrocephalus. J Fish Biol 77:1071–1082
Ouchi A, Hamasaki S (1979) Population analysis of the common mackerel, Scomber japonicus, based on the catch statistics and biological information in the western Japan Sea and the East China Sea. Bull Seikai Natl Fish Res Inst 53:125–152
Yan YM (1997) Biology of Pneumatophorus japonicus in Fujian offshore waters. Mar Fish 19:69–73 (in Chinese with English abstract)
Hsueh Y, Wang J, Chern CS (1992) The intrusion of the Kuroshio across the continental shelf northeast of Taiwan. J Geophys Res 97:14320–14323
Sun X (1987) Analysis of the surface path of the Kuroshio in the East China Sea. In: Sun X (ed) Essays on the investigation of Kuroshio. Ocean Press, Beijing, pp 1–14
Wu B (1982) Some problems on circulation study in Taiwan Strait. Taiwan Str 1:1–7
Acknowledgments
The present study could not have been carried out without the willing help of those listed below in collecting specimens: Mr. Xi Han, Mr. Dian-Rong Sun, Dr. Xiao Chen, Dr. Long-Shan Lin, Dr. Bo-Nian Shui. This study was supported by the International Cooperation and Exchange of the National Natural Science Foundation of China (No. 31061160187) and the Fundamental Research Funds for the Central Universities (No. 201262022).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, J., Yanagimoto, T., Song, N. et al. Population genetic structure of chub mackerel Scomber japonicus in the Northwestern Pacific inferred from microsatellite analysis. Mol Biol Rep 42, 373–382 (2015). https://doi.org/10.1007/s11033-014-3777-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3777-2