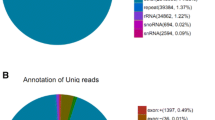
Abstract
Of late years, a large amount of conserved and species-specific microRNAs (miRNAs) have been performed on identification from species which are economically important but lack a full genome sequence. In this study, Solexa deep sequencing and cross-species miRNA microarray were used to detect miRNAs in white shrimp. We identified 239 conserved miRNAs, 14 miRNA* sequences and 20 novel miRNAs by bioinformatics analysis from 7,561,406 high-quality reads representing 325,370 distinct sequences. The all 20 novel miRNAs were species-specific in white shrimp and not homologous in other species. Using the conserved miRNAs from the miRBase database as a query set to search for homologs from shrimp expressed sequence tags (ESTs), 32 conserved computationally predicted miRNAs were discovered in shrimp. In addition, using microarray analysis in the shrimp fed with Panax ginseng polysaccharide complex, 151 conserved miRNAs were identified, 18 of which were significant up-expression, while 49 miRNAs were significant down-expression. In particular, qRT-PCR analysis was also performed for nine miRNAs in three shrimp tissues such as muscle, gill and hepatopancreas. Results showed that these miRNAs expression are tissue specific. Combining results of the three methods, we detected 20 novel and 394 conserved miRNAs. Verification with quantitative reverse transcription (qRT-PCR) and Northern blot showed a high confidentiality of data. The study provides the first comprehensive specific miRNA profile of white shrimp, which includes useful information for future investigations into the function of miRNAs in regulation of shrimp development and immunology.




Similar content being viewed by others
Abbreviations
- miRNAs:
-
MicroRNAs
- ESTs:
-
Expressed sequence tags
- qRT-PCR:
-
Quantitative real-time PCR
- miRNP:
-
miRNA ribonucleoprotein complex
- NCBI:
-
The National Center for Biotechnology Information
- snRNAs:
-
Small nuclear RNAs
- snoRNAs:
-
Small nucleolar RNAs
References
Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G et al (2009) Conserved microRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139:1096–1108
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Cui Q, Yu Z, Purisima EO, Wang E (2006) Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol 2:46
Friedman RC, Farh KKH, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360
Unver T, Namuth-Covert DM, Budak H (2009) Review of current methodological approaches for characterizing microRNAs in plants. Int J Plant Genomics. 2009:262463
Lu YZ, Liu J (2010) In silico identification of microRNAs and their targets in diatoms. Afr J Microbiol Res 4:1433–1439
Manila TM, Riju A, Lakshmi Priya Darshini K, Chandrasekar A, Eapen SJ (2009) In silico microRNA identification from paprika (Capsicum annuum) ESTs. Nature Precedings. http://hdl.handle.net/10101/npre.2009.3737.1
Zhang W, Luo Y, Gong X, Zeng W, Li S (2009) Computational identification of 48 potato microRNAs and their targets. Comput Biol Chem 33:84–93
Qiu CX, Xie FL, Zhu YY, Guo K, Huang SQ, Nie L et al (2007) Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene 395:49–61
Han YS, Luan FL, Zhu HL, Shao Y, Chen AJ, Lu CW et al (2009) Computational identification of microRNAs and their targets in wheat (Triticum aestivum L.). Science in China Series C. Life Sci 52:1091–1100
Sunkar R, Jagadeeswaran G (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8:37
Das A, Mondal TK (2010) Computational identification of conserved microRNAs and their targets in tea (Camellia sinensis). Am J Plant Sci 1(02):77
Tong C, Jin Y, Zhang Y (2006) Computational prediction of microRNA genes in silkworm genome. J Zhejiang University-Sci B. 7:806–816
Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z et al (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178
Yin Z, Shen F (2010) Identification and characterization of conserved microRNAs and their target genes in wheat (Triticum aestivum). Genet Mol Res 9:1186–1196
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK et al (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8:R96
Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C et al (2008) Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44:3–12
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25
Chen R, Hu Z, Zhang H (2009) Identification of microRNAs in wild soybean (Glycine soja). J Integr Plant Biol 51:1071–1079
Zhao CZ, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ et al (2010) Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol 10:3
Chang YS, Peng SE, Yu HT, Liu FC, Wang CH, Lo CF et al (2004) Genetic and phenotypic variations of isolates of shrimp Taura syndrome virus found in Penaeus monodon and Metapenaeus ensis in Taiwan. J Gen Virol 85:2963–2968
Yao X, Wang L, Song L, Zhang H, Dong C, Zhang Y et al (2010) A Dicer-1 gene from white shrimp Litopenaeus vannamei: expression pattern in the processes of immune response and larval development. Fish Shellfish Immunol 29:565–570
Labreuche Y, Veloso A, De La Vega E, Gross PS, Chapman RW, Browdy CL et al (2010) Non-specific activation of antiviral immunity and induction of RNA interference may engage the same pathway in the Pacific white leg shrimp Litopenaeus vannamei. Dev Comp Immunol 34:1209–1218
Liu XL, Xi QY, Yang L, Li HY, Jiang QY, Shu G et al (2011) The effect of dietary Panax ginseng polysaccharide extract on the immune responses in white shrimp Litopenaeus vannamei. Fish Shellfish Immunol 30:495–500
Li HY, Xi QY, Xiong YY, Liu XL, Cheng X, Shu G et al (2012) Identification and comparison of microRNAs from skeletal muscle and adipose tissues from two porcine breeds. Anim Genet 43:704–713
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Ruan L, Bian X, Ji Y, Li M, Li F, Yan X (2011) Isolation and identification of novel microRNAs from Marsupenaeus japonicus. Fish shellfish immunology 31(2):334–340
Willenbrock H, Salomon J, Søkilde R, Barken KB, Hansen TN, Nielsen FC et al (2009) Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA 15:2028–2034
Yousef M, Showe L, Showe M (2009) A study of microRNAs in silico and in vivo: bioinformatics approaches to microRNA discovery and target identification. FEBS J 276:2150–2156
Nasaruddin M, Harikrishna K, Othman Y, Hoon S, Harikrishna A (2007) Computational prediction of microRNAs from oil palm (Elaeis guineensis Jacq.) expressed sequence tags. Asian Pac J Mol Biol Biotechnol 15:107–113
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science’s STKE 294:858
Wei Y, Chen S, Yang P, Ma Z, Kang L (2009) Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol 10:R6
Okamura K, Phillips MD, Tyler DM, Duan H, Chou Y, Lai EC (2008) The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15:354–363
Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC (2008) The evolution and functional diversification of animal microRNA genes. Cell Res 18:985–996
Luo W, Sehgal A (2012) Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148:765–779
Acknowledgments
This study was supported by grants from the Chinese Key Basic Plan (No. 2011CB944200), and the Natural Science Foundation of Guangdong province (S2013010013215).
Conflicts of interest
The authors report no conflicts of interest with this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Qian-Yun Xi, Yuan-Yan Xiong and Yuan-Mei Wang: co-first authors and also contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xi, QY., Xiong, YY., Wang, YM. et al. Genome-wide discovery of novel and conserved microRNAs in white shrimp (Litopenaeus vannamei). Mol Biol Rep 42, 61–69 (2015). https://doi.org/10.1007/s11033-014-3740-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3740-2