Abstract
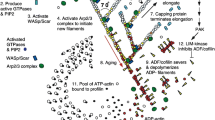
We report the development of a coarse-grained Langevin dynamics model of a lamellipodium featuring growing F-actin filaments in order to study the effect of stiffness of the F-actin filament, the G-actin monomer concentration, and the number of polymerization sites on lamellipodium protrusion. The virtual lamellipodium is modeled as a low-aspect-ratio doubly capped cylinder formed by triangulated particles on its surface. It is assumed that F-actin filaments are firmly attached to a lamellipodium surface where polymerization sites are located, and actin polymerization takes place by connecting a G-actin particle to a polymerization site and to the first particle of a growing F-actin filament. It is found that there is an optimal number of polymerization sites for rapid lamellipodium protrusion. The maximum speed of lamellipodium protrusion is related to competition between the number of polymerization sites and the number of available G-actin particles, and the degree of pulling and holding of the lamellipodium surface by non-polymerizing actin filaments. The lamellipodium protrusion by actin polymerization displays saltatory motion exhibiting pseudo-thermal equilibrium: the lamellipodium speed distribution is Maxwellian in two dimensions but the lamellipodium motion is biased so that the lamellipodium speed in the direction of the lamellipodium motion is much larger than that normal to the lamellipodium motion.
Similar content being viewed by others
References
Abraham, V.C., Krishnamurthi, V., Taylor, D.L., Lanni, F.: The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 77, 1721–1732 (1999)
Adelman, S.A., Doll, J.D.: Generalized Langevin equation approach for atom-solid-surface scattering—general formulation for classical scattering off harmonic solids. J. Chem. Phys. 64, 2375–2388 (1976)
Alberts, J.B., Odell, G.M.: In silico reconstitution of Listeria propulsion exhibits nano-saltation. Plos Biol. 2, 2054–2066 (2004)
Allen, M.P., Tildesley, D.J.: Computer Simulation of Liquids. Clarendon, Oxford (1987)
Atilgan, E., Wirtz, D., Sun, S.X.: Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 90, 65–76 (2006)
Bernheim-Groswasser, A., Wiesner, S., Golsteyn, R.M., Carlier, M.F., Sykes, C.: The dynamics of actin-based motility depend on surface parameters. Nature 417, 308–311 (2002)
Betz, T., Lim, D., Kas, J.A.: Neuronal growth: A bistable stochastic process. Phys. Rev. Lett. 96, 098103 (2006)
Bird, R.B., Curtiss, C.F., Armstrong, R.C., Hassager, O.: Dynamics of Polymeric Liquids: Kinetic Theory, 2nd edn. Wiley, New York (1987)
Bray, D.: Cell Movement. Garland, New York (1992)
Briels, W.J.: Theory of polymer dynamics. http://cbp.tnw.utwente.nl/PolymeerDictaat/polymerdynamics.pdf (1998)
Bryce, N.S., Clark, E.S., Leysath, J.L., Currie, J.D., Webb, D.J., Weaver, A.M.: Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15, 1276–1285 (2005)
Cascone, I., Audero, E., Giraudo, E., Napione, L., Maniero, F., Philips, M.R., Collard, J.G., Serini, G., Bussolino, F.: Tie-2-dependent activation of RhoA and Rac1 participates in endothelial cell motility triggered by angiopoietin-1. Blood 102, 2482–2490 (2003)
Claessens, M., Bathe, M., Frey, E., Bausch, A.R.: Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat. Mater. 5, 748–753 (2006)
Czirok, A., Schlett, K., Madarasz, E., Vicsek, T.: Exponential distribution of locomotion activity in cell cultures. Phys. Rev. Lett. 81, 3038–3041 (1998)
Dickinson, R.B., Purich, D.L.: Clamped-filament elongation model for actin-based motors. Biophys. J. 82, 605–617 (2002)
Dickinson, R.B., Tranquillo, R.T.: A stochastic-model for adhesion-mediated cell random motility and haptotaxis. J. Math. Biol. 31, 563–600 (1993)
Dimilla, P.A., Barbee, K., Lauffenburger, D.A.: Mathematical-model for the effects of adhesion and mechanics on cell-migration speed. Biophys. J. 60, 15–37 (1991)
Doi, M., Edwards, S.F.: The Theory of Polymer Dynamics. Oxford University Press, New York (1986)
Footer, M.J., Kerssemakers, J.W.J., Theriot, J.A., Dogterom, M.: Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl. Acad. Sci. USA 104, 2181–2186 (2007)
Frenkel, D., Smit, B.: Understanding Molecular Simulations: From Algorithms to Applications. Academic Press, New York (2002)
Gerbal, F., Chaikin, P., Rabin, Y., Prost, J.: An elastic analysis of Listeria monocytogenes propulsion. Biophys. J. 79, 2259–2275 (2000)
Gerbal, F., Laurent, V., Ott, A., Carlier, M.F., Chaikin, P., Prost, J.: Measurement of the elasticity of the actin tail of Listeria monocytogenes. Eur. Biophys. J. Biophys. 29, 134–140 (2000)
Giannone, G., Dubin-Thaler, B.J., Dobereiner, H.G., Kieffer, N., Bresnick, A.R., Sheetz, M.P.: Periodic lamellipodial contractions correlate with rearward actin waves. Cell 116, 431–443 (2004)
Gracheva, M.E., Othmer, H.G.: A continuum model of motility in ameboid cells. Bull. Math. Biol. 66, 167–193 (2004)
Herrmann, K.H., Satyanarayana, S.V.M., Sridhar, V., Murthy, K.P.N.: Monte Carlo simulation of actin filament based cell motility. Int. J. Mod. Phys. B 17, 5597–5611 (2003)
Isambert, H., Venier, P., Maggs, A.C., Fattoum, A., Kassab, R., Pantaloni, D., Carlier, M.F.: Flexibility of actin-filaments derived from thermal fluctuations—effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 (1995)
Jeon, J., Dobrynin, A.V.: Polymer confinement and bacterial gliding motility. Eur. Phys. J. E 17, 361–372 (2005)
Kuo, S.C., McGrath, J.L.: Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature 407, 1026–1029 (2000)
Mahadevan, L., Matsudaira, P.: Motility powered by supramolecular springs and ratchets. Science 288, 95–99 (2000)
Marcy, Y., Prost, J., Carlier, M.F., Sykes, C.: Forces generated during actin-based propulsion: A direct measurement by micromanipulation. Proc. Natl. Acad. Sci. USA 101, 5992–5997 (2004)
McBride, M.J.: Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55, 49–75 (2001)
Mogilner, A., Edelstein-Keshet, L.: Regulation of actin dynamics in rapidly moving cells: A quantitative analysis. Biophys. J. 83, 1237–1258 (2002)
Mogilner, A., Oster, G.: Force generation by actin polymerization II: The elastic ratchet and tethered filaments. Biophys. J. 84, 1591–1605 (2003)
Mombach, J.C.M., Glazier, J.A.: Single cell motion in aggregates of embryonic cells. Phys. Rev. Lett. 76, 3032–3035 (1996)
Moreno, J., Vargas, M.A., Madiedo, J.M., Munoz, J., Rivas, J., Guerrero, M.G.: Chemical and rheological properties of an extracellular polysaccharide produced by the cyanobacterium Anabaena sp ATCC 33047. Biotechnol. Bioeng. 67, 283–290 (2000)
Parekh, S.H., Chaudhuri, O., Theriot, J.A., Fletcher, D.A.: Loading history determines the velocity of actin-network growth. Nat. Cell Biol. 7, 1219–1223 (2005)
Peskin, C.S., Odell, G.M., Oster, G.F.: Cellular motions and thermal fluctuations—the Brownian ratchet. Biophys. J. 65, 316–324 (1993)
Pollard, T.D., Blanchoin, L., Mullins, R.D.: Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. 29, 545–576 (2000)
Prass, M., Jacobson, K., Mogilner, A., Radmacher, M.: Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 174, 767–772 (2006)
Rubinstein, B., Jacobson, K., Mogilner, A.: Multiscale two-dimensional modeling of a motile simple-shaped cell. Multiscale Model. Simul. 3, 413–439 (2005)
Satyanarayana, S.V.M., Baumgaertner, A.: Shape and motility of a model cell: A computational study. J. Chem. Phys. 121, 4255–4265 (2004)
Schaus, T.E., Taylor, E.W., Borisy, G.G.: Self-organization of actin filament orientation in the dendritic-nucleation/array-treadmilling model. Proc. Natl. Acad. Sci. USA 104, 7086–7091 (2007)
Stevens, M.J.: Simple simulations of DNA condensation. Biophys. J. 80, 130–139 (2001)
Theriot, J.A.: The polymerization motor. Traffic 1, 19–28 (2000)
Theriot, J.A., Mitchison, T.J., Tilney, L.G., Portnoy, D.A.: The rate of actin-based motility of intracellular listeria-monocytogenes equals the rate of actin polymerization. Nature 357, 257–260 (1992)
Zaman, M.H., Kamm, R.D., Matsudaira, P., Lauffenburger, D.A.: Computational model for cell migration in three-dimensional matrices. Biophys. J. 89, 1389–1397 (2005)
Zamir, E., Geiger, B.: Components of cell-matrix adhesions. J. Cell Sci. 114, 3577–3579 (2001)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeon, J., Alexander, N.R., Weaver, A.M. et al. Protrusion of a Virtual Model Lamellipodium by Actin Polymerization: A Coarse-Grained Langevin Dynamics Model. J Stat Phys 133, 79–100 (2008). https://doi.org/10.1007/s10955-008-9600-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10955-008-9600-5