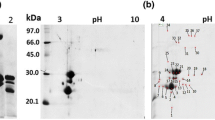
Abstract
CC-NBS-LRR (CNL) plant proteins are related with highly conserved family of disease resistance protein distinguished by a coiled-coil domain, which plays an important role in innate immunity. The present study reports the purification and identification of CNL like protein fragment (CNL-LPF) by two step chromatography and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF/MS), respectively. Furthermore, current study also illustrated the development of polyclonal antibody against purified CNL-LPF, which was used for immunolocalization of CNL-LPF in cytoplasm of cotyledon, using Fluorescence microscopy and Transmission electron microscopy. Lastly, present study also demonstrates in vitro oligomerization of purified CNL-LPF with multiple bands on 4–10 % gradient native-PAGE; each band representing a small fraction of each oligomer population as evident by immunoblots. In conclusion, the current study deals with the purification and polyclonal antibody development against CNL-LPF.







Similar content being viewed by others
Abbreviations
- CNL:
-
CC-NBS-LRR protein
- CNL-LPF:
-
CNL like protein fragment
- PAbs:
-
Polyclonal antibody
- ELISA:
-
Enzyme-linked immunosorbent assay
- FM:
-
Fluorescence microscopy
- TEM:
-
Transmission electron microscopy
- PM:
-
Plasma membrane
References
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR–encoding genes in arabidopsis. Plant Cell 15:809–834
Richly E, Kurth J, Leister D (2003) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19:76–84
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Głowacki S, Macioszek VK, Kononowicz AK (2011) R proteins as fundamentals of plant innate immunity. Cell Mol Biol Lett 16:1–24
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20:116–122
Leipe DD, Koonin EV, Aravind L (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 333:781–815
Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12:817–826
MacHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212
Bernoux M, Ellis JG, Dodds PN (2011) New insights in plant immunity signalling activation. Curr Opin Plant Biol 14:512–518
Inohara N, Chamaillard M, McDonald C, Nunez G (2005) NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem 74:355–383
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E (2009) Wheel of life, wheel of death: a mechanistic insight into signalling by STAND proteins. Structure 17:172–182
Venugopal SC, Jeong RD, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y (2009) Enhanced disease susceptibility1 and salicylic acid act redundantly to regulate resistance gene-mediated signalling. PloS Genet e1000545
Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B (2007) Nuclear activity of MLA immunereceptors links isolate- specific and basal disease-resistance responses. Science 315:1098–1103
Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22:4176–4194
Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1and is balanced by its functional domains. Plant Cell 22:4195–4215
Yadav S, Tomar A, Jithesh O, Khan M, Yadav R, Srinivasan A, Singh T, Yadav S (2011) Purification and partial characterization of low molecular weight vicilin-like glycoprotein from the seeds of citrullus lanatu. Protein J 30:575–580
Tarr DEK, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2:197
Drenckhahn D, Jöns T, Schmitz F (1993) Production of polyclonal antibodies against proteins and peptides. Methods Cell Biol 37:7–56
Grodzki AC, Berenstein E (2010) Antibody purification: affinity chromatography—protein A and protein G Sepharose. Methods Mol Biol 588:33–41
Pagnussat L, Burbach C, Baluška F, Canal L (2012) An extracellular lipid transfer protein is relocalized intracellularly during seed germination. J Exp Bot 63:6555–65563
Reyes D, Rodrı´guez D, Lorenzo O, Nicola´s G, Can˜as R, Canto´n FR, Canovas FM, Nicola´ C (2006) Immunolocalization of FsPK1 correlates this abscisic acid-induced protein kinase with germination arrest in Fagus sylvatica L. seeds. J Exp Bot 57:923–929
Jackson DP (1992) In situ hybridization in plants. In: Gurr SJ, McPhereson, Bowles DJ, eds. Molecular plant pathology: a practical approach 163–174
Barrasa JM, Gutiérrez A, Escaso V, Guillén F, Martínez MJ, Martínez AT (1998) Electron and fluorescence microscopy of extracellular glucan and aryl-alcohol oxidase during wheat-straw degradation by Pleurotus eryngii. Appl Environ Microbiol 64:325–332
Davis BJ (1964) Ann N Y Acad Sci 121:404–436
Scha¨gger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217:220–230
Piedras P, Rivas S, Dröge S, Hillmer S, Jones JDG (2000) Functional, c-myc-tagged Cf-9 resistance gene products are plasma-membrane localized and glycosylated. Plant J 21:529–536
Rivas S, Romeis T, Jones JDG (2002) The Cf-9 disease resistance protein is present in a ∼420-Kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14:689–702
Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158:1819–1832
Gu L, Guo R (2007) Genome-wide detection and analysis of alternative splicing for nucleotide binding site-leucine-rich repeats sequences in rice. J Genet Genom 34:247–257
Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AME, Rathjen JP (2010) Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J 61:507–518
Mestre P, Baulcombe DC (2006) Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18:491–501
Acknowledgments
This work is financially supported by Department of Science and Technology (DST), India) under the Women Scientists Scheme (WOS-A). Neha Kumari thanks DST for providing fellowship. We gratefully acknowledge Dr Radhey Shyam Sharma for fruitful discussion and Dr Kalpana Luthra for allowing us to use her laboratory facility during antibody development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
This article contain polyclonal antibody development on New Zealand white rabbits. This study was permitted by the AIIMS animal ethics committee (733/IAEC/13).
Rights and permissions
About this article
Cite this article
Kumari, N., Kumar, R., Mishra, V. et al. Polyclonal Antibody Development Against Purified CC-NBS-LRR like Protein Fragment from Mature Lageneria siceraria Seeds and Immunolocalization. Protein J 35, 379–390 (2016). https://doi.org/10.1007/s10930-016-9683-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-016-9683-9