Abstract
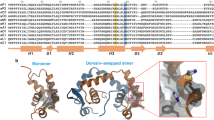
The forkhead box (FOX) proteins are a family of transcription factors that interact with DNA via a winged helix motif that forms part of the forkhead domain. The FOXP (FOXP1–4) subfamily is unique in the family in that the forkhead domains of these proteins are able to dimerise via domain swapping. In this event, structural elements are exchanged via extension of the hinge loop region. Despite the high sequence homology among the FOXP subfamily members, the stability of their forkhead domain dimers varies, with FOXP3 forming the most stable dimer. An amino acid difference is observed in the hinge region of the FOXP subfamily where a tyrosine in all members is replaced with a phenylalanine in FOXP3. In this work, the role of phenylalanine at this position in the hinge region was investigated. This was done by creating the Y540F variant of the FOXP2 forkhead domain. The effect of the Y540F mutation on the structure, dimerisation propensity and DNA binding ability of the FOXP subfamily was investigated. The mutation altered the structure of the protein by decreasing the disorder of the backbone as measured by circular dichroism spectroscopy and by altering the local environment of the hinge region as measured by tryptophan fluorescence. The propensity of the forkhead domain to form a dimer was improved ~9.5 fold by the mutation. This was attributed to increased hydrophobicity at the dimer interface as well as altered tension in the hinge loop region. DNA binding assays indicated that the affinity for DNA was decreased by the mutation. Taken together, these findings suggest that domain swapping may modulate DNA binding.





Similar content being viewed by others
Abbreviations
- CD:
-
Circular dichroism
- EMSA:
-
Electrophoretic mobility shift assay
- FHD:
-
Forkhead domain
- FOX:
-
Forkhead box family
- IPEX:
-
Immune dysregulation, polyendocrinopathy, enteropathy and X-linked
References
Kaufmann E, Knochel W (1996) Five years on the wings of fork head. Mech Dev 57(1):3–20
Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM (2003) Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 316:79–89
Chae WJ, Henegariu O, Lee SK, Bothwell AL (2006) The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc Natl Acad Sci USA 103(25):9631–9636
Su DM, Navarre S, Oh WJ, Condie BG, Manley NR (2003) A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat Immunol 4:1128–1135
Friedman J, Kaestner K (2006) The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci CMLS 63(19–20):2317–2328
Partridge L, Bruning JC (2008) Forkhead transcription factors and ageing. Oncogene 27(16):2351–2363
Lu MM, Li S, Yang H, Morrisey EE (2002) Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech Dev 119(Suppl 1):S197–S202
Rausa FM, Tan Y, Costa RH (2003) Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol Cell Biol 23(2):437–449
Mecklenburg L, Nakamura M, Sundberg JP, Paus R (2001) The nude mouse skin phenotype: the role of Foxn1 in hair follicle development and cycling. Exp Mol Pathol 71(2):171–178
Castanet M, Park SM, Smith A, Bost M, Leger J, Lyonnet S, Pelet A, Czernichow P, Chatterjee K, Polak M (2002) A novel loss-of-function mutation in TTF-2 is associated with congenital hypothyroidism, thyroid agenesis and cleft palate. Hum Mol Genet 11(17):2051–2059
Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA (2003) Structural and functional analyses of disease-causing missense mutations in the forkhead domain of FOXC1. Hum Mol Genet 12(22):2993–3005
Saleem RA, Banerjee-Basu S, Murphy TC, Baxevanis A, Walter MA (2004) Essential structural and functional determinants within the forkhead domain of FOXC1. Nucleic Acids Res 32(14):4182–4193
Hamdan FF, Daoud H, Rochefort D, Piton A, Gauthier J, Langlois M, Foomani G, Dobrzeniecka S, Krebs M-O, Joober R (2010) De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet 87(5):671–678
Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe A-M, Bird LE, Davies KE, Fisher SE (2006) Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet 15(21):3154–3167
Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD (2001) The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27(1):20–21
Bandukwala HS, Wu Y, Feuerer M, Chen Y, Barboza B, Ghosh S, Stroud JC, Benoist C, Mathis D, Rao A (2011) Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity 34(4):479–491
Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, Wood K, Cordell JL (2001) The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res 61(24):8820–8829
Wildin R, Smyk-Pearson S, Filipovich A (2002) Clinical and molecular features of the immunodysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet 39(8):537–545
Li S, Weidenfeld J, Morrisey EE (2004) Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24(2):809–822
Clark KL, Halay ED, Lai E, Burley SK (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5
van Dongen MJ, Cederberg A, Carlsson P, Enerbäck S, Wikström M (2000) Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11. J Mol Biol 296(2):351–359
Jin C, Marsden I, Chen X, Liao X (1999) Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol 289(4):683–690
Tsai KL, Huang CY, Chang CH, Sun YJ, Chuang WJ, Hsiao CD (2006) Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J Biol Chem 281(25):17400–17409
Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD (2007) Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res 35(20):6984–6994
Boura E, Rezabkova L, Brynda J, Obsilova V, Obsil T (2010) Structure of the human FOXO4-DBD-DNA complex at 1.9 a resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr Sect D Bioll Crystallogr 66(Pt 12):1351–1357
Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A (2010) Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res 38(13):4527–4538
Sheng W, Rance M, Liao X (2002) Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence. Biochemistry 41(10):3286–3293
Brent MM, Anand R, Marmorstein R (2008) Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16(9):1407–1416
Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L (2006) Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14(1):159–166
Schlunegger MP, Bennett MJ, Eisenberg D (1997) Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv Protein Chem 50:61–122
Liu Y, Gotte G, Libonati M, Eisenberg D (2002) Structures of the two 3D domain-swapped RNase A trimers. Protein Sci Publ Protein Soc 11(2):371–380
Rousseau F, Schymkowitz JW, Itzhaki LS (2003) The unfolding story of three-dimensional domain swapping. Structure 11(3):243–251
Park C, Raines RT (2000) Dimer formation by a “monomeric” protein. Protein Sci Publ Protein Soc 9(10):2026–2033
Janowski R, Kozak M, Jankowska E, Grzonka Z, Grubb A, Abrahamson M, Jaskolski M (2001) Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat Struct Biol 8(4):316–320
Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC (2001) Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol 8(9):770–774
Staniforth RA, Giannini S, Higgins LD, Conroy MJ, Hounslow AM, Jerala R, Craven CJ, Waltho JP (2001) Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily. EMBO J 20(17):4774–4781
D’Alessio G (1999) The evolutionary transition from monomeric to oligomeric proteins: tools, the environment, hypotheses. Prog Biophys Mol Biol 72(3):271–298
Gotte G, Bertoldi M, Libonati M (1999) Structural versatility of bovine ribonuclease A: distinct conformers of trimeric and tetrameric aggregates of the enzyme. Eur J Biochem FEBS 265(2):680–687
Liu Y, Eisenberg D (2002) 3D domain swapping: as domains continue to swap. Protein Sci Publ Protein Soc 11(6):1285–1299
Newcomer ME (2002) Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol 12(1):48–53
Cole N, Ralston GB (1994) Enhancement of self-association of human spectrin by polyethylene glycol. Int J Biochem 26(6):799–804
Lindner R, Ralston G (1995) Effects of dextran on the self-association of human spectrin. Biophys Chem 57(1):15–25
Rivas G, Fernandez JA, Minton AP (1999) Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry 38(29):9379–9388
Green SM, Gittis AG, Meeker AK, Lattman EE (1995) One-step evolution of a dimer from a monomeric protein. Nat Struct Mol Biol 2(9):746–751
Parker MJ, Dempsey CE, Hosszu LL, Waltho JP, Clarke AR (1998) Topology, sequence evolution and folding dynamics of an immunoglobulin domain. Nat Struct Mol Biol 5(3):194–198
Rousseau F, Schymkowitz JW, Wilkinson HR, Itzhaki LS (2001) Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc Natl Acad Sci USA 98(10):5596–5601
Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D (2001) Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc Natl Acad Sci 98(19):10687–10691
Seeliger MA, Schymkowitz JW, Rousseau F, Wilkinson HR, Itzhaki LS (2002) Folding and association of the human cell cycle regulatory proteins ckshs1 and ckshs2. Biochemistry 41(4):1202–1210
Byeon IJ, Louis JM, Gronenborn AM (2004) A captured folding intermediate involved in dimerization and domain-swapping of GB1. J Mol Biol 340(3):615–625
Frank MK, Dyda F, Dobrodumov A, Gronenborn AM (2002) Core mutations switch monomeric protein GB1 into an intertwined tetramer. Nat Struct Mol Biol 9(11):877–885
Chu YP, Chang CH, Shiu JH, Chang YT, Chen CY, Chuang WJ (2011) Solution structure and backbone dynamics of the DNA-binding domain of FOXP1: insight into its domain swapping and DNA binding. Protein Sci Publ Protein Soc 20(5):908–924
Whitmore L, Wallace BA (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89(5):392–400
Wang B, Lin D, Li C, Tucker P (2003) Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem 278(27):24259–24268
Correcirc DH, Ramos CH (2009) The use of circular dichroism spectroscopy to study protein folding, form and function. Afr J Biochem Res 3(5):164–173
Murray AJ, Lewis SJ, Barclay AN, Brady RL (1995) One sequence, two folds: a metastable structure of CD2. Proc Natl Acad Sci 92(16):7337–7341
Callis PR, Liu T (2004) Quantitative prediction of fluorescence quantum yields for tryptophan in proteins. J Phys Chem B 108(14):4248–4259
Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Acknowledgments
We extend our gratitude to Lin Chen, University of Southern California, Los Angeles, CA for the generous donation of the pET-30 LIC plasmid encoding FOXP2 FHD. This work was supported by the University of the Witwatersrand and the South African National Research Foundation Grants 80681, 89515 and 64788 from the South African Chairs Initiative of the Department of Science and Technology and National Research Foundation. Any opinion, findings and conclusions or recommendations expressed in this material are those of the author(s) and therefore the National Research Foundation, the Department of Science and Technology does not accept any liability with regard thereto.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Perumal, K., Dirr, H.W. & Fanucchi, S. A Single Amino Acid in the Hinge Loop Region of the FOXP Forkhead Domain is Significant for Dimerisation. Protein J 34, 111–121 (2015). https://doi.org/10.1007/s10930-015-9603-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-015-9603-4