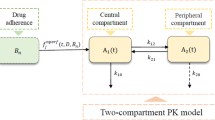
Abstract
Adherence is a frequent contributing factor to variations in drug concentrations and efficacy. The purpose of this work was to develop an integrated population model to describe variation in adherence, dose-timing deviations, overdosing and persistence to dosing regimens. The hybrid Markov chain–von Mises method for modeling adherence in individual subjects was extended to the population setting using a Bayesian approach. Four integrated population models for overall adherence, the two-state Markov chain transition parameters, dose-timing deviations, overdosing and persistence were formulated and critically compared. The Markov chain–Monte Carlo algorithm was used for identifying distribution parameters and for simulations. The model was challenged with medication event monitoring system data for 207 hypertension patients. The four Bayesian models demonstrated good mixing and convergence characteristics. The distributions of adherence, dose-timing deviations, overdosing and persistence were markedly non-normal and diverse. The models varied in complexity and the method used to incorporate inter-dependence with the preceding dose in the two-state Markov chain. The model that incorporated a cooperativity term for inter-dependence and a hyperbolic parameterization of the transition matrix probabilities was identified as the preferred model over the alternatives. The simulated probability densities from the model satisfactorily fit the observed probability distributions of adherence, dose-timing deviations, overdosing and persistence parameters in the sample patients. The model also adequately described the median and observed quartiles for these parameters. The Bayesian model for adherence provides a parsimonious, yet integrated, description of adherence in populations. It may find potential applications in clinical trial simulations and pharmacokinetic-pharmacodynamic modeling.



Similar content being viewed by others
References
Blaschke TF, Osterberg L, Vrijens B, Urquhart J (2012) Adherence to medications: insights arising from studies on the unreliable link between prescribed and actual drug dosing histories. Annu Rev Pharmacol Toxicol 52:275–301. doi:10.1146/annurev-pharmtox-011711-113247
Levy G (1993) A pharmacokinetic perspective on medicament noncompliance. Clin Pharmacol Ther 54(3):242–244
Levy G, Zamacona MK, Jusko WJ (2000) Developing compliance instructions for drug labeling. Clin Pharmacol Ther 68(6):586–591. doi:10.1067/mcp.2000.110976
Rubio A, Cox C, Weintraub M (1992) Prediction of diltiazem plasma concentration curves from limited measurements using compliance data. Clin Pharmacokinet 22(3):238–246
Vrijens B, Goetghebeur E (2004) Electronic monitoring of variation in drug intakes can reduce bias and improve precision in pharmacokinetic/pharmacodynamic population studies. Stat Med 23(4):531–544. doi:10.1002/sim.1619
Vrijens B, Goetghebeur E (1999) The impact of compliance in pharmacokinetic studies. Stat Methods Med Res 8(3):247–262
Savic RM, Barrail-Tran A, Duval X, Nembot G, Panhard X, Descamps D, Verstuyft C, Vrijens B, Taburet AM, Goujard C, Mentre F, Group ACS (2012) Effect of adherence as measured by MEMS, ritonavir boosting, and CYP3A5 genotype on atazanavir pharmacokinetics in treatment-naive HIV-infected patients. Clin Pharmacol Ther 92(5):575–583. doi:10.1038/clpt.2012.137
Kenna LA, Labbe L, Barrett JS, Pfister M (2005) Modeling and simulation of adherence: approaches and applications in therapeutics. AAPS J 7(2):E390–E407. doi:10.1208/aapsj070240
Feng Y, Gastonguay MR, Pollock BG, Frank E, Kepple GH, Bies RR (2011) Performance of Cpred/Cobs concentration ratios as a metric reflecting adherence to antidepressant drug therapy. Neuropsychiatr Dis Treat 7:117–125. doi:10.2147/NDT.S15921
Fellows K, Rodriguez-Cruz V, Covelli J, Droopad A, Alexander S, Ramanathan M (2015) A hybrid Markov chain–von Mises density model for the drug-dosing interval and drug holiday distributions. AAPS J 17(2):427–437. doi:10.1208/s12248-014-9713-5
Rajarshi MB (2012) Markov chains and their extensions. Statistical inference for discrete time stochastic processes. Springer, New York. doi:10.1007/978-81-322-0763-4_2
Devore J (1976) A note on the estimation of parameters in a Bernoulli model with dependence. Ann Stat 4(5):990–992. doi:10.1214/aos/1176343597
Rudolfer SM (1990) A Markov chain model of extrabinomial variation. Biometrika 77(2):255–264
Islam MN, O’shaughnessy CD (2013) On the Markov chain binomial model. Appl Math 4:1726–1730
Wong D, Modi R, Ramanathan M (2003) Assessment of Markov-dependent stochastic models for drug administration compliance. Clin Pharmacokinet 42(2):193–204. doi:10.2165/00003088-200342020-00006
Agostinelli A, Lund U (2013) Circular statistics (version 0.4-7). Department of Environmental Sciences, Informatics and Statistics, Foscari University, Venice
Weibull model: problem with slow mixing and effective sample (2015). http://sourceforge.net/p/mcmc-jags/discussion/610036/thread/d5249e71/
Team RC (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Plummer M, Stukalov A, Denwood M (2015) Bayesian graphical models using MCMC: interface to the JAGS MCMC library. http://mcmc-jags.sourceforge.net
Plummer M, Best N, Cowles K, Vines K (2006) CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11
Brooks SP, Gelman A (1998) General methods for monitoring convergence of iterative simulations. J Comput Graph Stat 7:434–455
Gelman A, Carlin JB, Stern HS, Rubin DB (2004) Bayesian data analysis. Texts in statistical science. CRC Press, Boca Raton
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–511
Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A (2002) Bayesian measures of model complexity and fit (with discussion). JR Stat Soc B 64(4):583–639. doi:10.1111/1467-9868.00353
Plummer M (2008) Penalized loss functions for Bayesian model comparison. Biostatistics 9(3):523–539. doi:10.1093/biostatistics/kxm049
Fellingham GW, Kottas Athanasios, Hartman BM (2015) Bayesian nonparametric predictive modeling of group health claims. Insurance 60:1–10
Müller P, Quintana F (2004) Nonparametric Bayesian data analysis. Stat Sci 19(1):95–110
Girard P, Blaschke TF, Kastrissios H, Sheiner LB (1998) A Markov mixed effect regression model for drug compliance. Stat Med 17(20):2313–2333
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer, New York
Zhu L, Carlin BP (2000) Comparing hierarchical models for spatio-temporally misaligned data using the deviance information criterion. Stat Med 19(17–18):2265–2278
Acknowledgments
This paper is a tribute and acknowledgement of the numerous seminal contributions of Dr. Gerhard Levy to the pharmaceutical sciences. My research interests in adherence modeling were sparked by an incidental conversation with Dr. Gerhard Levy in the corridor many years ago. We take this opportunity to congratulate and felicitate Dr. Levy on his 50-year track record of scientific accomplishments. We are grateful to Colin Stoneking and Klaus Oberauer (Department of Cognitive Psychology, University of Zurich, Switzerland) for kindly providing the add-on code for analyzing the VM distribution. This research is not funded. Support from the National Multiple Sclerosis Society (RG4836-A-5) to the Ramanathan laboratory is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fellows, K., Stoneking, C.J. & Ramanathan, M. Bayesian population modeling of drug dosing adherence. J Pharmacokinet Pharmacodyn 42, 515–525 (2015). https://doi.org/10.1007/s10928-015-9439-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-015-9439-8