Abstract
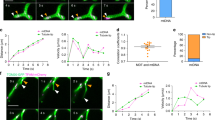
Mitochondrial nucleoids are confined sites of mitochondrial DNA existing in complex clusters with the DNA–compacting mitochondrial (mt) transcription factor A (TFAM) and other accessory proteins and gene expression machinery proteins, such as a mt single–stranded–DNA–binding protein (mtSSB). To visualize nucleoid distribution within the mt reticular network, we have employed three–dimensional (3D) double–color 4Pi microscopy. The mt network was visualized in hepatocellular carcinoma HepG2 cells via mt–matrix–addressed GFP, while 3D immunocytochemistry of mtSSB was performed. Optimization of iso-surface computation threshold for nucleoid 4Pi images to 30 led to an average nucleoid diameter of 219 ± 110 and 224 ± 100 nm in glucose– and galactose–cultivated HepG2 cells (the latter with obligatory oxidative phosphorylation). We have positioned mtDNA nucleoids within the mt reticulum network and refined our model for nucleoid redistribution within the fragmented network –– clustering of up to ten nucleoids in 2 μm diameter mitochondrial spheroids of a fragmented mt network, arising from an original 10 μm mt tubule of a 400 nm diameter. However, the theoretically fragmented bulk parts were observed most frequently as being reintegrated into the continuous mt network in 4Pi images. Since the predicted nucleoid counts within the bulk parts corresponded to the model, we conclude that fragmentation/reintegration cycles are not accompanied by mtDNA degradation or that mtDNA degradation is equally balanced by mtDNA replication.







Similar content being viewed by others
References
Bahlmann K, Jakobs S, Hell SW (2001) 4Pi-confocal microscopy of live cells. Ultramicroscopy 87:155–164
Bogenhagen DF, Rousseau D, Burke S (2008) The layered structure of human mitochondrial DNA nucleoids. J Biol Chem 283:3665–3675
Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA (2011) Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol Cell Biol 31:4994–5010
Cree LM, Samuels DC, Lopes S, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HHM, Chinnery PF (2008) A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet 40:249–254
Dlasková A, Špaček T, Šantorová J, Plecitá-Hlavatá L, Berková Z, Saudek F, Lessard M, Bewersdorf J, Ježek P (2010) 4Pi microscopy reveals an impaired three-dimensional mitochondrial network of pancreatic islet beta-cells in an experimental model of type-2 diabetes. Biochim Biophys Acta Bioenerg 1797:1327–1341
Egner A, Jakobs S, Hell SW (2002) Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc Natl Acad Sci U S A 99:3370–3375
Gilkerson RW, Schon EA, Hernandez E, Davidson MM (2008) Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol 181:1117–1128
Gomes LC, Benedetto GD, Scorrano L (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13:589–598
Gugel H, Bewersdorf J, Jakobs S, Engelhardt J, Storz R, Hell SW (2004) Cooperative 4Pi excitation and detection yields sevenfold sharper optical sections in live-cell microscopy. Biophys J 87:4146–4152
Hell SW, Stelzer EHK (1992a) Properties of a 4Pi-confocal fluorescence microscope. J Opt Soc Am A 9:2159–2166
Hell SW, Stelzer EHK (1992b) Fundamental improvement of resolution with a 4Pi-confocal fluorescence microscope using two-photon excitation. Opt Commun 93:277–282
Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN (2007) Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion 7:311–321
Ježek P, Plecitá-Hlavatá L (2009) Mitochondrial reticulum network dynamics in relation to oxidative stress, redox regulation, and hypoxia. Int J Biochem Cell Biol 41:1790–1804
Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF (2012) Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci U S A 109:6136–6141
Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jacobs S (2011) Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A 108:13534–13539
Mlodzianoski MJ, Schreiner JM, Callahan SP, Smolková K, Dlasková A, Šantorová J, Ježek P, Bewersdorf J (2011) Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt Express 19:15009–15019
Plecitá-Hlavatá L, Lessard M, Šantorová J, Bewersdorf J, Ježek P (2008) Mitochondrial oxidative phosphorylation and energetic status are reflected by morphology of mitochondrial network in INS-1E and HEP-G2 cells viewed by 4Pi microscopy. Biochim Biophys Acta 1777:834–846
Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64:985–993
Spelbrink JN (2010) Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62:19–32
Stewart JB, Larsson NG (2014) Keeping mtDNA in shape between generations. PLoS Genet 10:e1004670
Stewart JB, Treter C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG (2008) Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol 6:e10
Tauber J, Dlasková A, Šantorová J, Smolková K, Alán L, Špaček T, Plecitá-Hlavatá L, Jabůrek M, Ježek P (2013) Distribution of mitochondrial nucleoids upon mitochondrial network fragmentation and network reintegration in HEPG2 cells. Int J Biochem Cell Biol 45:593–603
Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27:433–446
Twig G, Liu X, Liesa M, Wikstrom JD, Molina AJ, Las G, Yaniv G, Hajnóczky G, Shirihai OS (2010) Biophysical properties of mitochondrial fusion events in pancreatic beta-cells and cardiac cells unravel potential control mechanisms of its selectivity. Am J Physiol Cell Physiol 299:C477–C487
Wang Y, Bogenhagen DF (2006) Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem 281:25791–25802
Acknowledgments
The project was supported by grants of the Grant Agency of the Czech Republic (GACR) No. 13-02033S to P.J., and No. P305/12/1247 to M.J.; by the research projects AV0Z50110509 and RVO67985823 to the Institute of Physiology; and also by the grant project BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University“ (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund and by the grant project The Centre of Biomedical Research (CZ.1.07/2.3.00/30.0025). The two latter sources were also co-funded by the European Social Fund and the state budget of the Czech Republic.
Compliance with ethical standards
All authors declare no conflict of interest. This work represents the new work concerning an expansion of the previous work cited in the References.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dlasková, A., Engstová, H., Plecitá – Hlavatá, L. et al. Distribution of mitochondrial DNA nucleoids inside the linear tubules vs. bulk parts of mitochondrial network as visualized by 4Pi microscopy. J Bioenerg Biomembr 47, 255–263 (2015). https://doi.org/10.1007/s10863-015-9610-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-015-9610-3