Abstract
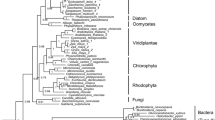
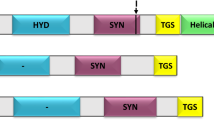
Uridine diphosphate (UDP)-glucose pyrophosphorylase (UGPase) is an essential enzyme in algae, catalyzing the phosphorylation that forms UDP-glucose, the key precursor for biosynthesis of carbohydrates such as trehalose, cellulose, and starch. However, the current knowledge about algal UGP is quite little. Here, we conducted transcriptomic and genomic sequence analyses for algal UGPs. Only one UGP sequence was predicted in each red and brown algal species, and a particular UGP/phosphoglucomutase (PGM) fusion gene was found in some brown algae. Sequence comparisons indicated that these predicted UGPs exhibited significant identity to their fungal and plant orthologs, and the fusion UGP/PGM also had active sites conserved in eukaryotes. Phylogenetic analysis suggested that eukaryotic algal and plant UGPs may originate from non-cyanobacterial prokaryotes via horizontal gene transfer, and the non-fusion UGP is the ancestral form. During the evolution of brown algae, UGP had undergone gene duplication and merged with a PGM segment, leading to UGP/PGM. This duplication may not exist in the common ancestor of brown algae and diatoms, but possibly occurred after species differentiation. To study the biochemical properties of algal UGPase, Gracilaria chouae UGP (GcUGP), Saccharina japonica UGP (SjUGP), and S. japonica UGP/PGM (SjUGP/PGM) were recombinantly expressed. GcUGP and SjUGP proteins catalyzed the phosphorylation reaction, with GcUGP having a higher catalytic efficiency (k cat /K m ) than SjUGP, and the optimal reaction conditions differed partly between these two species, possibly due to their divergent developmental processes and adaptabilities to marine environment. However, little UGPase or PGM activities were detected in fusion protein, which indicates the gene duplication may lead to gene neofunctionalization. The results offer a theoretical basis for further study of the role of UGPase in algae and algal functional product synthesis pathways.





Similar content being viewed by others
References
Abe T, Niiyama H, Sasahara T (2002) Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor Appl Genet 105:216–221
Ap Rees T (1998) Hexose phosphate metabolism by nonphotosynthetic tissues of higher plants. Biochem Plants 14:1–33
Armbrust EV, Berges JA, Bowler C et al (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Arturo OL, Mark AR (1998) Characterization of the UDP-glucose pyrophosphorylase gene from the marine red alga Gracilaria gracilis. J Appl Phycol 10:581–588
Bowler C, Allen AE, Badger JH et al (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Chang LP, Sui ZH, Fu F, Zhou W, Wang JG, Kang KH, Zhang S, Ma JH (2014) Relationship between gene expression of UDP-glucose pyrophosphorylase and agar yield in Gracilariopsis lemaneiformis (Rhodophyta). J Appl Phycol 26:2435–2441
Coleman HD, Ellis DD, Gilbert M, Mansfield SD (2006) Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotech J 4:87–101
Coleman HD, Canam T, Kang KY, Ellis DD, Mansfield SD (2007) Over-expression of UDP-glucose pyrophosphorylase in hybrid poplar affects carbon allocation. Exp Bot 58:4257–4268
Daniel D, Meng M, Agnieszka G, Anders H, Malgorzata W, Leszek AK (2012) Substrate kinetics and substrate effects on the quaternary structure of barely UDP-glucose pyrophosphorylase. Phytochemistry 79:39–45
Diez-Roux G, Banfi S, Sultan M et al (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9(1):e1000582
Elling L, Kula MR (1994) Purification of UDP-glucose pyrophosphorylase from germinated barley (malt). J Biotechnol 34:157–163
Geisler M, Wilczynska M, Karpinski S, Kleczkowski LA (2004) Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: homology-modeling analyses. Plant Mol Biol 56:783–794
Herman MK, Beatriz B, Agnete MP (1953) Uridy1 transferases and the formation of uridine triphosphate: uridy1 transferases and the formation of uridine diphosphogalactose. Nature 172:1036–1037
Huelsenbeck JP, Ronquist FR (2001) MrBayes: Bayesian inference of phylogeny. Bioformatics 17:754–755
Ingeborg CB, Ana R, Michiel K, Willem MV (2001) Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesus. Appl Environ Microbiol 67:3033–3040
Jean MD, Nathalie D, Denise TS, Veronique P, Jean F (1953) Genetic and biochemical characterization of the UGP1 gene encoding the UDP-Glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem 233:520–530
Kim JS, Koh S, Shin HJ, Lee DS, Lee SY (1999) Biochemical characterization of a UDP-sugar pyrophosphorylase from Thermus caldophilus GK24. Biotechnol Appl Biochem 29:11–17
Kim MS, Kim M, Terada R, Yang EC, Boo SM (2008) Gracilaria parvispora is the correct name of the species known as G. bursa-pastoris in Korea and Japan. Taxon 57:231–237
Lack AR, Robert PD (1987) Structure and sequence of a UDP glucose pyrophosphorylase gene of Dictyostelium discoideum. Nucl Acids Res 15:3891–3906
Lai XH, Wu J, Chen S, Zhang XL, Wang HH (2008) Expression, purification, and characterization of a functionally active Mycobacterium tuberculosis UDP-glucose pyrophosphorylase. Protein Expr Purif 61:50–56
Leszek AK (1994) Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry 37:1507–1515
Li TY, Ren L, Zhou G, Liu C, Chi S, Liu T (2012) A suitable method for extracting total RNA from red algae. Trans Oceanol Limnol 4:63–71
Lu X, Li MZ, Xu ZG, Wang XY (2013) Impact of light intensity on growth and levels of photosynthetic pigments of Gracilaria chouae. Progr Fish Sci 34:145–150 (in chinese)
Ma Z, Fan HJ, Lu CP (2011) Molecular cloning and analysis of the UDP-glucose pyrophosphorylase in Streptococcus equi subsp. zooepidemicus. Mol Biol Rep 38:2751–2760
Marques AR, Ferreira PB, Correia IS, Fialho AM (2003) Characterization of the ugpG gene encoding a UDP-glucose pyrophosphorylase from the gellan gum producer Sphingomonas paucimobilis ATCC31461. Mol Genet Genomics 268:816–824
Meng M, Malgorzata W, Leszek AK (2008) Molecular and kinetic characterization of two UDP-glucose pyrophosphorylases, products of distinct genes, from Arabidopsis. BBA-Proteins Proteom 1784:967–972
Meng M, Fitzek E, Gajowniczek A, Wilczynska M, Kleczkowski LA (2009) Domain-specific determinants of catalysis/structure binding and the oligomerization status of barley UDP-glucose pyrophosphorylase. BBA-Proteins Proteom 1794:1734–1742
Michel G, Tonon T, Scornet D, Cock JM, Kloareg B (2010) Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: insights into the origin and evolution of storage carbohydrates in Eukaryotes. New Phytol 188:67–81
Munch-Petersen A, Kalckar HM, Cutolo E, Smith EE (1953) Uridyl transferases and the formation of uridine triphosphate; enzymic production of uridine triphosphate: uridine diphosphoglucose pyrophosphorolysis. Nature 172:1036–1037
Noriyasu O, Saori Y, Naoko T, Chizu K, Yoshitsugu S, Koji T (2008) Escherichia coil cytosolic glycerophosphodiester phosphodiesterase (UgpQ) requires Mg2+, Co2+, or Mn2+ for its enzyme activity. J Bacteriol 4:1219–1223
Page RD (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Peng HL, Chang HY (1993) Cloning of a human liver UDP-glucose pyrophosphorylase cDNA by complementation of the bacterial galU mutation. FEBS Lett 329:153–158
Posada D (2003) Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinforma. doi:10.1002/04471250953.bio0605s00
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Qiu H, Yoon HS, Bhattacharya D (2013) Algal endosymbionts as vectors of horizontal gene transfer in photosynthetic eukaryotes. Front Plant Sci 19(4):366
Rastogi S, Liberles DA (2005) Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol 5(1):28
Ronquist F, Huelsenbeck JP (2003) Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Shaikh AH, Katsuyuki T, Yasuaki K, Toshio F (1994) Overproduction and characterization of recombinant UDP-Glucose pyrophosphorylase from Escherichia coli K-12. J Nutr Biochem 115:965–972
Su JC, Hassid WZ (1962) Carbohydrates and nucleotides in the red alga Porphyra perforata II. separation and identification of nucleotides. Biochemistry 1:474–480
Sun YY, Luo D, Zhao C, Li W, Liu T (2011) DNA extraction and PCR analysis of five kinds of large seaweed under different preservation conditions. Mol Plant Breeding 9:1680–1691
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang QH, Zhang X, Li FG, Hou YX, Liu XL, Zhang XY (2011) Identification of a UDP-glucose pyrophosphorylase from cotton (Gossypium hirsutum L.) involved in cellulose biosynthesis in Arabidopsis thaliana. Plant Cell Rep 30:1303–1312
Wickens GE (2001) Useful Algae. Economic Botany. Springer, Netherlands, pp 373–388
Yoshihiro K, Ratsuyuki T, Susumu M, Toshio F (1993) Molecular cloning, nucleotide sequencing, and affinity labeling of bovine liver UDP-Glucose pyrophosphorylase. J Biotechnol 114:61–68
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China under contract nos. 41206116 and 41376143.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
The nucleotide and deduced amino acid sequences of GcUGP (A), SjUGP (B), and SjUGP/PGM (C), and schematic drawings of the recombinant plasmids GcUGP/pET32a (D), SjUGP/pET32a (E), SjUGP/PGM/pET32a (F) (JPEG 8 kb)
Figure S2
SDS-PAGE analysis of purified fusion proteins from Escherichia coli cells. Lane M: Prestained protein molecular mass marker. Lane 1: G. chouae UGP with a calculated molecular mass of 72 KDa. Lane 2: S. japonica UGP with a calculated molecular mass of 71 KDa (JPEG 29 kb)
ESM 1
(DOC 87 kb)
ESM 2
(DOC 32 kb)
Rights and permissions
About this article
Cite this article
Chi, S., Feng, YJ. & Liu, T. Molecular cloning, characterization, and comparison of UDP-glucose pyrophosphorylase from Gracilaria chouae and Saccharina japonica . J Appl Phycol 28, 2051–2059 (2016). https://doi.org/10.1007/s10811-015-0738-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-015-0738-7