Abstract
Climate change accelerates biodiversity alterations in northern ecosystems. A prevalent example is that tundra regions are invaded by boreal species. This impacts negatively on native species through competition, predation and transmission of zoonoses. Scandinavian red foxes (Vulpes vulpes) have emerged into the tundra and have altered the structure and function of the tundra community. For instance, they threaten persistence of the endangered Swedish Arctic fox (Vulpes lagopus). County board administrations implement control of the tundra red foxes, but little is known about the underlying expansion dynamics. A broad-scale study revealed high connectivity where northern areas were supplemented with red foxes from surrounding population. However, red fox expansion is most prevalent in tundra regions and the fine-scaled expansion dynamics in these areas have not yet been disseminated. With the aim of identifying the invasive pathways of tundra red foxes, we present microsatellite data for 205 Swedish red foxes and mitochondrial sequence variation in 102 foxes sampled across the historical boreo-nemoral distribution and recently colonized tundra regions. Genetic structuring was low with high levels of ongoing, asymmetric dispersal from surrounding boreal zones into tundra habitats causing high genetic admixture. In both tundra and boreo-nemoral regions, inter-individual relatedness decreased with increasing geographic distance and data suggests male-biased dispersal patterns. Overall, fine-scaled expansion patterns were affected by multiple factors and we discuss its implications for future red fox management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ongoing climate change is forecasted to reduce and fragment mountain tundra habitats and induce alterations in the ecological function and dynamics (Guisan et al. 2014; Elmhagen et al. 2015). One consequence of the milder climate is that boreal species expand from southern sources and establish in northern habitats, a process that by many means is comparable to that of invasive species taking over novel habitats. There are two, not mutually exclusive processes, underlying this process (Elmhagen et al. 2015). In southern areas, warmer temperatures and increased land use induce demographic growth that in combination with range expansion will increase the distribution and abundance of boreal species. Meanwhile, the opposite occurs in northern habitats, where reductions in habitat availability cause population declines and/or range contractions of populations living under sub-Arctic conditions (Elmhagen et al. 2015). These declines can be further accelerated by boreal species advancing into tundra regions and change ecosystem dynamics by competition or predation (Ims and Fuglei 2005), or by introducing novel diseases and parasites (e.g. Kutz et al. 2005). There is thus a prevalent need to devote research focus towards the boreal expansion process to implement efficient and ethical management strategies as well as for conservation of threatened native species (Elmhagen et al. 2015).
Efforts to limit expanding or invasive species have often been impeded by lack of relevant information related to the species ecology and demography (Bomford and O’Brien 1995). For successful eradication, for example, it is necessary for removal rates to exceed population growth and that the population is isolated from immigration (Bomford and O’Brien 1995; Harding et al. 2001). Clearly identifying the invasive pathways and source population is crucial to avoid devoting effort towards populations that are likely to become recolonized in a short time perspective (Robertson and Gemmel 2004; Estoup and Guillemaud 2010). Including genetic analysis as a tool can thus supplement management decisions with information about the recent and historical origin, composition and connectivity of expanding populations (Estoup and Guillemaud 2010). In addition to this, modern analytical approaches allow for recent migration routes and dispersal rates to be disseminated (Andreasen et al. 2012). Hence, in order to formulate realistic management approaches and to predict the future impact of invasions, it is necessary to retrieve detailed information about the fine-scaled expansion patterns and dispersal routes of highly influential and expanding species (Rollins et al. 2009; Estoup and Guillemaud 2010).
The red fox (Vulpes vulpes) is a classic example of a species undergoing major population expansions. As a consequence of human-mediated landscape changes and increasing temperatures in various ecosystems, the European red fox population has during the last century expanded in numbers and geographic range (Hersteinsson and Macdonald 1992; Selås and Vik 2006; Elmhagen and Rushton 2007). In southern Sweden, the red fox population underwent a demographic increase in response to the intensified land use during the nineteenth century (Elmhagen and Rushton 2007), while red foxes in northern areas expanded in range during the same time period (Hersteinsson and Macdonald 1992). Tundra red fox abundance may benefit from changes in reindeer herding strategies that mediates higher food availability (Killengreen et al. 2011). Numerous climatic, landscape and biological factors have been suggested to influence red fox dispersal behavior. For instance, European red fox abundance and dispersal is limited by winter harshness (Hersteinsson and Macdonald 1992; Selås and Vik 2006; Barton and Zalewski 2007; Pasanen-Mortensen et al. 2013). Furthermore, the decision to disperse and establish in a novel habitat may be further influenced by sex-specific dispersal patterns and/or social factors (Lawson-Handley and Perrin 2007). A preference for establishing in territories adjacent to close relatives, or synchronize dispersal with close relatives have been documented in a closely related species (Ehrich et al. 2012; Elmhagen et al. 2015). It is however not known how these factors influence to the ongoing red fox invasion of the Fennoscandian tundra, but it is possible that there may be a higher occurrence of close relatives in the expansion front.
Increases and establishment of red foxes in tundra regions have affected the vulnerable tundra ecosystem in several ways (Macpherson 1964; Hersteinsson and Macdonald 1992). For example, the red fox has contributed to alterations in community structure and declines of competitor and prey populations (Hersteinsson and Macdonald, 1992; Smedshaug et al. 1999; Angerbjörn et al. 2013). In particular, the emergence of red foxes into the tundra region is associated with Arctic fox (Vulpes lagopus) population decline and withdrawal into areas of lower quality (Dalén et al. 2004; Angerbjörn et al. 2013). The patterns observed in these sympatric fox populations is an example of the expansion–contraction scenarios outlined by Elmhagen et al. (2015). Furthermore, the red fox is a host and vector for parasites such as sarcoptic mange (Sarcoptes scabiei) and fox tapeworm (Echinococcus multilocularis) as well as contagious and lethal diseases like arctic rabies (Letkova et al. 2006). Immigration and establishment of red foxes on the tundra thus convey considerable risks of introducing and transmitting these diseases and parasites to native species (Berteaux et al. 2004).
To minimize the ecological consequences and to conserve the critically endangered arctic fox in Sweden, there is a need to manage the expanding tundra populations of red foxes. Previous work has demonstrated that the negative effects exerted by red foxes on arctic foxes can be reduced through intensive culling in specific core areas (Angerbjörn et al. 2013). Swedish authorities have included red fox removal as a vital action in the management plan for Scandinavian arctic foxes (Elmhagen 2008). The efficiency of red fox culling however varies considerably across different areas and counties (Angerbjörn et al. 2013). These actions, however, are both expensive and time-consuming and the practical challenges for working in tundra areas put high demands on well-planned logistics, experienced personnel and large sources of funding. To accomplish efficient and ethical control of tundra red foxes it is necessary to define target areas of specific demographic importance, which requires understanding dispersal routes.
We previously conducted a broad-scale study on the expansion dynamics across Fennoscandia and found that northern areas were supplemented with immigrants from several surrounding regions (Norén et al. 2015). However, the detailed process underlying red fox emergence and establishment in tundra areas was not specifically addressed. In this paper, we investigate their origin and relationship between tundra and boreal red foxes. We used 15 highly variable microsatellite loci and one mitochondrial fragment (cytochrome-b, 354 bp) to investigate (i) genetic structuring between historical distribution (boreo-nemoral areas) and recently colonized (tundra) areas; (ii) the genetic composition and fine-scaled structure of the recently colonized tundra regions.
Methods
Sampling and genetic analyses
We assembled 53 red fox samples collected from the tundra regions in Sweden (62°55′N–68°77′N) between 2009 and 2013. Of these, 32 individuals were included in a previously published analysis of the large-scaled expansion patterns in Fennoscandia (Norén et al. 2015). The tundra foxes were obtained as tissue samples from red foxes culled for management purposes (procedure described by Angerbjörn et al. 2013) (n = 32) or faecal samples collected at known arctic and red fox den sites during Arctic fox inventories (n = 21) (Johansson et al. 2009). For comparison, we included the genotypes of 152 previously published red foxes (Norén et al. 2015). All samples were scored for variation in 15 autosomal (AHT140, c01.424PET, FH2004, FH2010, FH2088, FH2289, FH2380, AHT133, RF08.618, RF2001Fam, RF2054, CXX-468, CXX-602, FH2848 and REN54P11) and one sex-specific microsatellite (K9-Amelo) loci (one allele located on the Y chromosome and the other on the X chromosome) following Moore et al. (2010). Primer sequences for all microsatellite loci are provided in Moore et al. (2010) and references therein. All individual genotypes are available in Appendix in supplementary material. The K9-Amelo locus was used to identify the sex in faecal samples and to verify the visually identified sex for culled red foxes. Among the visually sex-determined foxes that had been collected from culling, the accuracy of the genetic sex determination was 100%.
For mitochondrial analysis, we amplified a cytochrome-b (354 bp) fragment in the mitochondria in 102 individuals sampled throughout Sweden using primers RF14724 (5′-CAACTATAAGAACATTAATGACC-3′) and RF15149 (5′-CTCAGAATGATATTTGTCCTC-3′) (Perrine et al. 2007; Aubry et al. 2009) on a PTC100 Programmable Thermal Controller (MJ Research Inc.). Each PCR setup was accompanied by negative controls from the extraction as well as PCR blanks. PCR products were checked through 1.5% agarose gel electrophoresis and thereafter cleaned using the PCR Purification Kit (Qiagen) or the ExoSAP-IT (Affymetrix) according to the manufacturer’s instructions. For all individuals, we sequenced the 5′ portion of the cytochrome-b fragment at the commercial lab Macrogen Inc. (www.macrogen.com). Sequences were verified for accurate species origin using BLAST, controlled visually and aligned in BioEdit 7.2.5 and haplotypes were determined against previously published data (Statham et al. 2014). Novel haplotypes (n = 6) were deposited on GenBank (Accession No KX768845-50). To verify the accuracy of our results and to avoid sequencing errors, we made an additional PCR reaction and thereafter sequenced the 3′ portion of each novel haplotype (i.e. previously unpublished ones).
Population structure
We used a Bayesian clustering approach as implemented in the software STRUCTURE 2.3.4 (Pritchard et al. 2009) to evaluate the occurrence of a spatial structure in the full sample of red foxes (tundra, boreal and nemoral zones; n = 205) as well as within the tundra zone (n = 53). For both data sets, we used the admixture model with correlated allele frequencies and 100 000 burn-in steps followed by one million MCMC replicates. We investigated the likelihood for K = 1–6 and each K value was repeated ten times. We determined the most likely number of K, using Structure Harvester (Earl and vonHoldt 2012) to calculate which value of K that displayed the highest L(K) and ΔK (Evanno et al. 2005). We thereafter merged the runs for the most likely value of K using CLUMPP (Jakobsson and Rosenberg 2007). Individual assignment over the averaged runs (obtained from CLUMPP) was accomplished using a threshold value of q > 0.80. Thereafter, we repeated the same STRUCTURE analysis using the LOCPRIOR model (Pritchard et al. 2009). We constructed a grid with cell size covering 20 × 20 km2. The cell size corresponds to a scale where gene flow still can occur between neighboring cells (Larvière and Pasitschniak-Arts 1996). Our data set subsequently consisted of 18 cells with a sample size of 4–26 individuals. Each individual was assigned a prior location, based on which grid cell they had been sampled in. We used 100 000 burn-in generations followed by one million MCMC replicates and ran the LOCPRIOR model for K = 1–6.
Levels of differentiation between the tundra and boreo-nemoral regions was quantified through population pairwise FST with significance testing using 10 000 permutations in Arlequin version 3.5.1.2 (Excoffier et al. 2005). We inferred levels of genetic admixture in the boreo-nemoral region as well as the recently colonized tundra region by implementing the USEPOPINFO option in STRUCTURE Pritchard et al. 2009). We used the admixture model with correlated allele frequencies and 100 000 burn-in steps followed by 106 MCMC replicates for K = 2 (tundra vs. boreal-nemoral) and GENSBACK = 0. We repeated the simulations three times, using a migration rate of v = 0.05. To test for differences in population membership values (q) among runs, i.e. the posterior probability of an individual originating from the same population where it was sampled, we used a non-parametric Kruskal–Wallis test in R (Sacks et al. 2005).
Distribution of genetic diversity
We tested for Hardy–Weinberg and linkage disequilibrium in tundra versus boreal-nemoral zones using Arlequin 3.5 (Excoffier et al. 2005). Hardy–Weinberg proportions were calculated using a Markov chain based on 10,000 dememorization steps while we used ten initial conditions to test for linkage disequilibrium. Significance testing was accomplished using 10,000 permutations and were corrected for multiple testing using the sequential Bonferroni correction (Holm 1979). To avoid sample size biases, we calculated allelic richness in FSTAT 2.9.3.2 (Goudet et al. 2002). Since it is theoretically expected that recently colonized areas display lower genetic variation through leading edge colonization and founder effects (Excoffier and Ray 2008), we tested for cluster-level difference in levels of heterozygosity and allelic richness using a Kruskal–Wallis test in R. Areas that recently have been colonized from a southern source are expected to display a south-to-north gradient in genetic variation (Norén et al. 2015) We tested for such a gradient in the level of genetic variation on the tundra, using the relationship between individual multi-locus heterozygosity (MLH) and latitude through a linear regression test in R.
We used the grid cell set-up to test for presence of isolation by distance between grid cells (described above) using a Mantel test based on linearized FST values and average sampling coordinates (Rousset 1997). To compare levels of admixture between cells, we calculated cell-specific FIS values and tested for a latitudinal effect on average FIS as well as the proportion of significantly deviant loci through linear regressions in R.
For mitochondrial DNA, we used Arlequin version 3.5.1.2 (Excoffier et al. 2005) to calculate haplotype and nucleotide diversity within each region as well as FST between regions. For significance testing, we used 10,000 permutations. We used the same software to calculate a minimum spanning network that was visualized in HapStar 0.7 (Teacher and Griffiths 2011). For reference and alignment, we used 53 cytochrome-b haplotypes previously published by Statham et al. (2014) and references therein.
Estimating recent dispersal rates
To test how red foxes in tundra and boreal regions were connected, we simulated the dispersal rates between tundra region grid cells and their neighboring boreal cells over the last generation using the software BIMr 1.1 (Faubet and Gaggiotto 2008). This is a recommended approach for populations connected by high ongoing gene flow since the ‘F model’ accepts deviations from genetic equilibrium (Faubet and Gaggiotto 2008; Andreasen et al. 2012. We implemented simulations with the two data sets, each containing one tundra cell (20 × 20 km) with a sample size of n > 10 individuals (termed Helags and Borgafjall respectively) and all neighboring boreal cells (termed Boreal1-5). We used the default settings in the software and, for each data set, we repeated the simulation three times. After checking for consistency between the three replicates, we thereafter chose the run displaying lowest Bayesian deviance, measured by the assignment values (D assign ) (Faubet and Gaggiotto 2008). For each area, we identified significant asymmetric dispersal through comparison of the 95% high density predictive interval (HDPI).
Determinants of genetic structure
We used the software GESTE 2.0 (Foll and Gaggiotti 2006) to identify population structure determinants using a Bayesian approach. The FST values for each grid cell are calculated and the effect of environmental factors versus genetic drift on genetic divergence was evaluated using a generalized linear model for the grid cells (described above). We evaluated signatures of range expansion, demographic and geographic heterogeneity as well as winter severity (Hersteinsson and Macdonald 1992; Selås and Vik 2006; Barton and Zalewski 2007; Pasanen-Mortensen et al. 2013) by comparing three different models; (i) the ‘range expansion model’, where the average sampling coordinates for each grid were used as explanatory factors, (ii) the ‘bioclimatic zone—abundance model’, where the bioclimatic zone and estimated population densities (assembled from Pasanen-Mortensen et al. 2013) for each cell were used as explanatory factors, and (iii) the ‘temperature—snow cover model’ where mean January temperature (Tveito et al. 2000) and yearly mean snow cover duration in days between 2000 and 2011 (Dietz et al. 2012) were assembled to approximate winter severity. For all simulations, we used the GESTE 2.0 default options and repeated each model three times to check for consistency between the runs.
We tested for the impact of geographic distance on inter-individual divergence through spatial autocorrelation tests in the software GenAlEx 6.41 (Peakall and Smouse 2006) using a distance class size of 100 km and significance testing using 9999 permutations. Tests were implemented in the full data set as well as for males and females separately. To further assess the occurrence of sex-specific dispersal patterns among tundra and boreo-nemoral foxes respectively, we implemented a frequency-based population assignment test (Paetkau et al. 1999) in the software GeneClass2 (Piry et al. 2004) for each sex separately (based on the genetic sex identification). For the dispersing sex, a lower mean assignment index (mAIC) and higher variance in the assignment indices (vAIC) is expected (Goudet et al. 2002; Podgorski et al. 2014). We tested from differences in mean assignment indices (mAIC) between males and females using a non-parametric Kruskal–Wallis test in R. In addition to this, we calculated maximum likelihood-based relatedness between all individuals in the software ML Relate 1.0 (Kalinowski et al. 2006). We also tested for the impact of geographic distance and pairwise relatedness for the full data set as well as males and females separately through a Mantel test with 999 permutations.
Results
Genetic structure and diversity
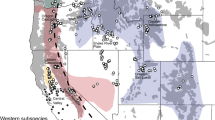
Based on the STRUCTURE analyses, K = 2 displayed highest L(K) and deltaK values (Fig. 1a, b). Among the tundra red foxes, 24.5% were assigned to the southerly cluster (blue in Figs. 1c, 2a), 52.8% to the northerly cluster (red in Figs. 1c, 2a) and 22.6% were un-assigned (q < 0.8). For foxes sampled in the boreal region, similar proportions were assigned to each cluster (35.6 and 37.3% respectively), while foxes from the nemoral region mainly were assigned to the southerly cluster (63.8 and 17% respectively). The LOCPRIOR model produced comparable clustering results for K = 2 (Fig. 1d). Genetic differentiation between the two regions was significant, but low (FST = 0.018, P < 0.01). The level of heterozygote deficiencies were comparable between the areas (sequential Bonferroni corrected α = 0.004 and 0.006 for tundra and boreo-nemoral regions respectively; Table 1) while there was no or low linkage disequilibrium between loci (Bonferroni corrected α = 0.00047; Table 1). Average heterozygosity and allelic richness was comparable between the areas (Table 1).
a Geographic distribution of individuals with population membership values of q > 0.8 in a specific cluster (shown in blue and red color) and unassigned individuals (in white) for K = 2 without prior population information over five replicates generated from CLUMPP. The dashed line represent Limes norrlandicus (i.e. the climate transition between boreo-nemoral and boreal habitats), b direction of significant, asymmetric dispersal over the last generation as inferred by BIMr where grey areas represent the tundra zones. (Color figure online)
For the mitochondrial data, we documented a total of nine different haplotypes. Among these, four were present in the tundra region and all nine in the boreo-nemoral region (Table 1). The previously documented haplotypes, U and U4 has a Eurasian distribution whereas haplotype U66 only has been documented in Sweden (Table 1, Statham et al. 2014). The remaining six haplotypes were previously unpublished (GenBank Accession nos KX768845–KX768850) and were closely related (separated by single substitutions) to the U, U4 or U66 haplotypes (Table 1; Fig. 3). As can be seen from the minimum spanning network, there were no unique tundra haplotypes, while five haplotypes were present only in the boreo-nemoral zone as and four were shared across all regions (Fig. 3). Gene diversity was 0.585 in the tundra and 0.709 in the boreo-nemoral region and nucleotide diversity ranged between 0.0018 and 0.0021. Genetic divergence between regions was non-significant (F ST = − 0.015, P = 0.621).
Minimum spanning network for the 354 base pair mitochondrial fragment cytochrome-b in the 102 Swedish red foxes. Orange color represent haplotypes shared across tundra, boreal and nemoral zones, blue color represent haplotypes only present in boreo-nemoral regions and white color represent haplotypes sampled in potential source populations, but not in Sweden (collected from Statham et al. (2014) and references therein). Haplotype frequency is represented by the size of the haplotype. (Color figure online)
Dispersal and connectivity
Based on the USEPOPINFO approach in STRUCTURE, red foxes sampled in tundra areas displayed more variable and significantly lower population membership values (q) compared to foxes sampled in nemoral and boreal areas (Kruskal–Wallis X2 = 5.67, df = 1, P = 0.017) (Fig. 4). Furthermore, with an assumed migration rate of v = 0.05, six first generation immigrants from boreal and nemoral areas were identified in tundra areas while only two immigrants from the tundra were identified in boreal and nemoral areas.
There was no significant effect of isolation by distance between cells in the grid (Mantel test: r xy = −0.017, P = 0.42) which suggests that additional factors influence population structure. Also, no apparent latitudinal effect on average FIS values (indicating admixture) (r 2 = −0.062, P = 0.959) or the proportion of significantly deviant loci (r 2 = 0.004, P = 0.318) was identified. Individual multi-locus heterozygosity in tundra regions ranged between 0.154–1, but there was no latitudinal effect on the distribution of heterozygosity (r2 = −0.018, P = 0.788).
Estimates of recent dispersal rates in the software BIMr revealed significant asymmetric dispersal patterns where immigration from neighboring boreal cells into the tundra regions of Helags and Borgafjäll were significantly higher compared to dispersal in the opposite direction (Table 2; Fig. 2b).This implies that tundra regions are supplemented by immigrants from neighboring boreal zones. In the GESTE simulations, we tried to disentangle which factors that influenced the dispersal patterns and genetic divergence (Table 3). Results for each model were comparable for all three replicates. For the ‘range expansion model’, highest posterior probability was recorded for an interaction between latitude and longitude (Pr = 0.839) (Table 2). When including genetic drift into the model, highest support was recorded for a scenario that combined effects of drift and spatial coordinates (Pr = 0.839) (Table 2). This means that in addition to genetic drift, there is an impact of dispersal in multiple directions that influences genetic divergence. For the ‘bioclimatic—population density model’ and ‘temperature—snow cover model’, the constant factor received highest posterior probability (Table 2). This means that genetic drift was likely to have a higher impact on genetic structuring than bioclimatic zone, population density, temperature or snow cover.
The spatial autocorrelation tests showed no significance of geographic distance regardless of using the full data set or for the tundra and boreo-nemoral data sets separately (P > 0.05). Also, testing males and females separately did not affect the outcome of the results. The assignment indices displayed comparable variance between males and females. The average assignment index was however significantly lower for tundra males than tundra females (Kruskal–Wallis X2 = 4.39, df = 1, P = 0.036), which is in agreement with the expectations for the dispersing sex (i.e. lower mAIC). For boreo-nemoral foxes, however, there was no difference (Kruskal–Wallis X2 = 0.08, df = 1, P = 0.78). Also, there was no difference in average relatedness between males (r = 0.047 ± 0.136) and females (r = 0.030 ± 0.078). The corresponding values among boreo-nemoral foxes were r = 0.06 ± 0.135 for males and r = 0.07 ± 0.133 for females. The distribution of relatedness was significantly correlated to geographic distance for the full data set (r = −0.264, P = 0.001) and males (r = −0.334, P = 0.002), whereas females only displayed a trend towards the same pattern (r = −0.156, P = 0.063). In the boreo-nemoral zone, there was similar patterns with a significant correlation between geographic and genetic distance for males (r = −0.173, P = 0.001) and close to significance for females (r = −0.074, P = 0.066).
Discussion
Expansion of boreal species is forecasted to become one of the most influential processes influencing northern biodiversity. The overall aim of this paper was to elucidate the fine-scaled genetic relationship between Swedish red foxes in recently colonized tundra areas and red foxes in the boreal and nemoral zones with specific emphasis on identifying the invasive pathways. Based on the mitochondrial DNA, the historical origin of tundra red foxes could not be elucidated since there was little divergence and all documented haplotypes (except the novel Swedish ones) are common and widespread across Eurasia (Statham et al. 2014). These results were in agreement with a general pattern of low mitochondrial structuring across large parts of the European mainland (Statham et al. 2014). The U and U4 haplotypes belong to the Holarctic clade whereas the U66 haplotype only has been identified in Sweden (Statham et al. 2014). The minimum spanning network illustrates the high level of haplotype sharing across bio-climatic zones and also the close relationship (one-base mutations) to the central and widespread U and U4 haplotypes (Fig. 3). The novel haplotypes originate from central, high-frequency haplotypes (U and U4) which is the pattern expected in a population undergoing expansion. This, in combination, with the lack of unique tundra haplotypes, suggest a common historical origin and lack of historical isolation between red foxes in the tundra and the rest of Sweden.
Despite the common historical origin, there may still be more recent divergence reflected in the microsatellite structure. The significant variance in population membership values between areas and high number of first-generation immigrants however confirm the previously documented pattern of high immigration into northern areas (Norén et al. 2015). Further, major deviations from genetic equilibrium indicate high levels of admixture. Interestingly, we also detected heterozygote deficiencies among red foxes in the boreo-nemoral zone. This finding suggests that there are high levels of ongoing gene flow in this region, which also is evident from the low levels of genetic divergence inferred by STRUCTURE (Fig. 1). With regards to the origin of expansion, there was low support for a single southern source of the tundra red foxes. Apart from a lower mitochondrial gene diversity on the tundra, our data do not support a simple scenario of northwards range expansion, which would be in agreement with previous broad-scale findings (Norén et al. 2015). The GESTE ‘range expansion model’ with combined effects of latitude, longitude and genetic drift received highest support, implying an expansion in multiple directions and from several (Table 3). Hence, the surrounding boreal areas are of high biological relevance by producing an excess of red foxes that emigrate and establish on the tundra.
Males and females displayed no difference in average relatedness, but there was a stronger effect of geographic distance on the distribution of male relatives where relatedness decreased with increasing spatial distance for both tundra and boreo-nemoral foxes. For tundra foxes, this was in concordance with the significant difference in assignment indices where red fox males displayed lower mean assignment index which indicates higher dispersal frequencies. For boreo-nemoral foxes, however, no such pattern was identified. Although this needs further evaluation, this data suggest that male-biased dispersal patterns may contribute to colonization and establishment on the tundra. Levels of relatedness may influence dispersal decisions and boreal red foxes may thus be more prone to emerge to the tundra if following a close relative. In the closely related arctic fox, presence of cohort-specific kin structure has been identified (Ehrich et al. 2012) and synchronized long distance dispersal by closely related, young males has been recorded (unpublished data). Male-biased kin structures have been documented in canid species such as the Ethiopian wolf (Canis simiensis; Gottelli et al. 2013) and, under specific demographic conditions, in the bat-eared fox (Otocyon megalotis; Kamler et al. 2013).
Implications for red fox management and arctic fox conservation
The low levels of genetic structuring in the Swedish red fox is likely a consequence of high dispersal across the entire distribution. Dispersal patterns were asymmetric where boreal zones supplement tundra areas with immigrants to a higher extent than the opposite. Our results bring important implications for the future management of northern red foxes. Based on the conclusions from this study, the tundra red fox population is supplemented with immigrants from the edge of the boreal zone. This means that efforts to control the tundra red fox population should be extended to also include the adjacent boreal zones. Otherwise, it is a prevalent risk that the tundra will be rapidly recolonized from the boreal forest (e.g. Robertson and Gemmel 2004). Furthermore, the finding of high connectivity between the boreal zone and the tundra emphasizes the importance of conducting red fox management as a permanent action. Also, to increase the efficiency of this action as well as avoiding unmotivated culling, an evaluation of the spatial scale where measures are necessary is highly recommended. Even though predator control has been successful in the case of Arctic fox conservation (Angerbjörn et al. 2013), there are examples of unexpected consequences of predator removal (Ruscoe et al. 2011). Therefore, continuous assessment of removal strategies and consequences is central.
This paper also highlights the competence of genetic analysis as a tool for wildlife management. For elusive and highly mobile species, such as the red fox, there are no direct approaches of elucidating dispersal patterns on a landscape scale. By taking advantage of the genetic signatures created by dispersal, a larger understanding of the process underlying expansion and invasions can be retrieved (Estoup and Guillemaud 2010). Since population expansions are predicted to become even more powerful processes during future decades as temperatures increase, we recommend that genetic tools should be incorporated as a vital part of the management of boreal, expanding populations with the aim of understanding, controlling and predicting their impact of northern ecosystems.
References
Andreasen AM, Stewart KM, Longland WS, Beckmann JP, Forister ML (2012) Identification of source sink dynamics in mountain lions of the Great Basin. Mol Ecol 21:5689–5701
Angerbjörn A, Eide NE, Dalén L et al (2013) Carnivore conservation in practice: replicated management actions on a large spatial scale. J Appl Ecol 50:59–67
Aubry KB, Statham MJ, Sacks BN et al (2009) Phylogeography of the North American red fox: vicariance in Pleistscene forest refugia. Mol Ecol 18:2668–2686
Barton KA, Zalewski A (2007) Winter severity limits red fox populations in Eurasia. Glob Ecol Biogeogr 16:281–289
Berteaux D, Reale D, McAdam SG et al (2004) Keeping pace with fast climate change: can Arctic life count on evolution? Integr Comp Biol 44:140–151
Bomford M, O’Brien P (1995) Eradication or control for vertebrate pests. Wildl Soc Bull 23:249–255
Dietz AJ, Wohner C, Kuenzer C (2012) European snow cover characteristics between 2000 and 2011 derived from improved MODIS daily snow cover products. Remote Sens 4:2432–2454
Dalén L, Elmhagen B, Angerbjörn A (2004) DNA analysis on fox faeces and competition induced niche shifts. Mol Ecol 13:2389–2392
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359
Ehrich D, Carmichael L, Fuglei E (2012) Age-dependent genetic structure of arctic foxes in Svalbard. Polar Biol 35:53–62
Elmhagen B, Kindberg J, Hellström P et al (2015) A boreal invasion in response of climate change? Range shifts and community effects in the borderland between forest and tundra. Ambio 44:39–50
Elmhagen B (2008) Åtgärdsprogram för fjällräv 2008–2012. Action plan, Swedish Environmental Protection Agency. Report 5927
Elmhagen B, Rushton SP (2007) Trophic control of mesopredators in terrestrial ecosystems: top-down or bottom-up? Ecol Lett 10:197–206
Estoup A, Guillemaud T (2010) Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol 19:4113–4130
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolut Bioinform Online 1:47–50
Excoffier L, Ray N (2008) Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol 23:347–351
Faubet P, Gaggiotto OE (2008) A new Bayesian method to identify the environmental factors that influence recent migration. Genetics 178:1491–1504
Foll M, Gaggiotti OE (2006) Identifying the environmental factors that determine the genetic structure of populations. Genetics 174:875–891
Gottelli D, Zillero-Subiri C, Marino J, Funk SM, Wang J (2013) Genetic structure and patterns of gene flow among populations of the endangered Ethiopian wolf. Anim Conserv 16:234–247
Goudet J (1995) FSTAT version 1.2: a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal usinig bi-parentally inherited genetic markers. Mol Ecol 11:1103–1114
Guisan A, Petitpierre B, Broenninmann O et al (2014) Unifying niche shift studies: insights from biological invasions. Trends Ecol Evol 29:260–268
Harding EK, Doak DF, Albertson JD (2001) Evaluating the effectiveness of predator control: the non-native red fox as a case study. Conserv Biol 15:1114–1122
Hersteinsson P, Macdonald DW (1992) Interspecific competition and the geographical distribution of red and arctic foxes Vulpes vulpes and Alopex lagopus. Oikos 64:505–515
Holm S (1979) A simple sequential rejective method procedure. Scand J Stat 6:65–70
Ims RA, Fuglei E (2005) Trophic interaction cycles in tundra ecosystems and the impact of climate change. Bioscience 55:311–322
Jakobsson M, Rosenberg N (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Johansson L, Mangi AC, Norén K, Angerbjörn A (2009) Spillnings inventering av fjällräv i Norrbottens län 2009. Länsstyrelsens Rapportserie 7:2009
Kalinowski ST, Wagner AP, Taper ML (2006) ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6:576–579
Kamler JM, Gray MM, Oh A, Macdonald DW (2013) Genetic structure, spatial organization, and dispersal in two populations of bat-eared foxes. Ecol Evol 3:2892–2902
Killengreen ST, Lecomte N, Ehrich D, Schott T, Yoccoz NG, Ims RA (2011) The importance of marine vs. human induced subsidies in the maintenance of an invasive mesocarnivore in the Arctic tundra. J Anim Ecol 80:10494–11060
Kutz SJ, Hoberg EP, Polley L, Jenkin EJ (2005) Global warming is changing the dynamics of Arctic host-parasite systems. Proc R Soc 272:2571–2576
Larvière S, Pasitschniak-Arts M (1996) Vulpes Vulpes. Mamm Species 537:1–11
Lawson-Handley L, Perrin N (2007) Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol 16:1559–1578
Letkova V, Lazar P, Curlik J et al (2006) The red fox (Vulpes vulpes L.) as a source of zoonoses. Vet Arch 76:73–81
Macpherson AH (1964) A northward range extension of the red fox in the eastern Canadian arctic. J Mamm 45:138–140
Norén K, Statham MJ, Ågren EO et al (2015) Genetic footprints reveal geographic patterns of expansion in Fennoscandian red foxes. Glob Change Biol 21:3299–3312
Moore M, Brown SK, Sacks BN (2010) Thirty-one short red fox (Vulpes vulpes) microsatellite markers. Mol Ecol Resour 10:404–408
Pasanen-Mortensen M, Pyykonen M, Elmhagen B (2013) Where lynx prevail, foxes will fail—limitation of a mesopredator in Eurasia. Glob Ecol Biogeogr 22:868–877
Paetkau D, Amstrup SC, Born EW et al (1999) Genetic structure of the world’s polar bear populations. Mol Ecol 8:1571–1584
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrine JD, Pollinger JP, Sacks BN et al (2007) Genetic evidence for the persistence of the critically endangered Sierra Nevada red fox in California. Conserv Gen 8:1083–1095
Piry S, Alapetite A, Cornuet JM et al (2004) GeneClass2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Wen X, Falush D (2009) STRUCTURE ver. 2.3. University of Chicago, Chicago. http://pritch.bsd.uchicago.edu/
Podgorski T, Scandura M, Jedrzejewska B (2014) Next of kin next door—philopatry and socio-genetic population structure in wild boar. J Zool 294:190–197
Robertson BC, Gemmel NJ (2004) Defining eradication units to control invasive pests. J Appl Ecol 41:1042–1048
Rollins LA, Woolnough AP, Wilton AN et al (2009) Invasive species can’t cover their tracks: using microsatellites to assist management of starling (Sturnus vulgaris) populations in Western Australia. Mol Ecol 18:1560–1573
Rousset F (1997) Genetic differentiation and estimation of gene flow from F statistics under isolation by distance. Genetics 145:1219–1228
Ruscoe WA, Ramsey DSL, Pech RP, Sweetapple PJ, Yockney I, Barron MC, Perry M, Nugent G, Carran R, Warne R, Brausch C, Duncan RP (2011) Unexpected consequences of control: competitive vs. predator release in a four-species assemblage of invasive mammals. Ecol Lett 14:1035–1042
Sacks BN, Mitchell BR, Williams CL et al (2005) Coyote movements and social structure along a cryptic population genetic subdivision. Mol Ecol 14:1241–1249
Selås V, Vik JO (2006) Possible impact of snow depth and ungulate carcasses on red fox (Vulpes vulpes) populations in Norway, 1897–1976. J Zool 269:299–308
Smedshaug CA, Selås V, Lund SE et al (1999) The effect of a natural reduction of red fox Vulpes vulpes on small game hunting bags in Norway. Wildl Biol 5:157–166
Statham MJ, Murdoch J, Janecka J et al (2014) Range-wide multilocus phylogeography of the red fox reveals ancient continental divergence, minimal genomic exchange, and distinct demographic histories. Mol Ecol 23:4813–4830
Teacher AGF, Griffiths DJ (2011) HapStar: automated haplotype network layout and visualization. Mol Ecol Resour 11:151–153
Tveito OE, Forland E, Heino R, Hanssen-Bauer I, Alexandersson H, Dahlström B, Drebs A, Kern-Hansen C, Jónsson T, Vaarby Laursen E, Westman Y (2000) Nordic temperature maps. DNMI report 09/00 KLIMA
Acknowledgements
We are grateful to rangers at the county board administrations in Jämtland, Västerbotten and Norrbotten, to E. O. Ågren at the National Veterinary Institute, Uppsala, Sweden and to T. and L. Norén, J.O. Andersson, N. Larsson, B. Lundström, S. Sorsén and M. Thorfve for collecting red fox samples and to M.J. Statham for help in the DNA lab. The study received financial support from Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) (#2011-1015 and #2010-263), the Göran Gustafsson foundation for Nature and Environment in Lappland, the foundation in memory of Oscar and Lili Lamm and the Tullberg foundation for biological research to KN.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2016_910_MOESM1_ESM.xlsx
Appendix 1. Geographical origin and individual multi-locus genotypes for each sample and primer information (Moore et al. 2010) for the 15 microsatellite loci used in this paper (XLSX 38 kb)
Appendix 1 Geographical origin and individual multi-locus genotypes for each sample and primer information (Moore et al. 2010) for the 15 microsatellite loci used in this paper
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Norén, K., Angerbjörn, A., Wallén, J. et al. Red foxes colonizing the tundra: genetic analysis as a tool for population management. Conserv Genet 18, 359–370 (2017). https://doi.org/10.1007/s10592-016-0910-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-016-0910-x