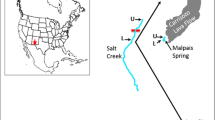
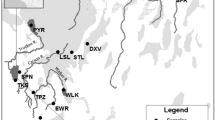
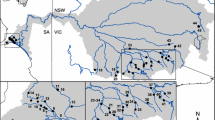
Abstract
Conservation genetic studies are challenged by the fact that populations of many imperiled species have experienced declines and fragmentation to the degree they no longer exhibit natural, self-sustaining metapopulation processes; characteristics of great importance to managers charged with their protection. Genetic patterns of species from minimally impacted systems can inform management practices for populations in more modified and fragmented systems. We assessed spatial and temporal patterns of intraspecific genetic diversity and differentiation using microsatellites for three imperiled fishes of the unfragmented upper Gila River, New Mexico, USA. Estimates of contemporary effective size were low for these species, but we observed little genetic evidence of inbreeding. Overall genetic structure was low (all species FST < 0.025) suggesting moderate to high gene flow for all species, but each exhibited different patterns of spatial structuring. Gila nigra (a candidate for listing under the Endangered Species Act) appears most at risk of short-term loss of genetic variation and local extinction relative to Meda fulgida or Rhinichthys (Tiaroga) cobitis (both federally endangered) because G. nigra exhibited the lowest diversity, smallest effective size (Ne ~100) and temporally unstable population structure. Meda fulgida and R. cobitis exhibited temporally stable spatial structure related to riverscape features but connectivity among occupied habitats is threatened by a proposed diversion structure. Data from this comparatively pristine system can inform management of these species in fragmented portions of their ranges.




Similar content being viewed by others
References
Allan JD, Flecker AS (1993) Biodiversity conservation in running waters. Bioscience 43:32–43
Apodaca JJ, Trexler JC, Jue NK, Schrader M, Travis J (2013) Large-scale natural disturbance alters genetic population structure of the sailfin molly, Poecilia latipinna. Am Nat 181:254–263
Blakney JR, Loxterman JL, Keeley ER (2014) Range-wide comparisons of northern leatherside chub populations reveal historical and contemporary patterns of genetic variation. Conserv Genet 15:757–770
Carlson CA, Muth RT (1989) The Colorado River: lifeline of the American Southwest. Can Spec Publ Fish Aquat Sci 106:220–239
Crispo E, Chapman LJ (2010) Temporal variation in population genetic structure of a riverine African cichlid fish. J Hered 101:97–106
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fagan WF (2002) Connectivity, fragmentation, and extinction risk in dendritic metapopulations. Ecology 83:3243–3249
Fagan WF, Unmack PJ, Burgess C, Minckley WL (2002) Rarity, fragmentation, and extinction risk in desert fishes. Ecology 83:3250–3256
Finn DS, Blouin MS, Lytle DA (2007) Population genetic structure reveals terrestrial affinities for a headwater stream insect. Freshw Biol 52:1881–1897
Fradkin PL (1981) A river no more: the Colorado River and the West. University of Arizona Press, Tucson
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R, Ballou JD, Briscoe DA (2009) Introduction to conservation genetics, 2nd edn. Cambridge University Press, New York
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Fraser DF, Gilliam JF, Yiphoi T (1995) Predation as an agent of population fragmentation in a tropical watershed. Ecology 76:1461–1472
Fraser DJ, Debes PV, Bernatchez L, Hutchings JA (2014) Population size, habitat fragmentation, and the nature of adaptive variation in a stream fish. Proc R Soc B Biol Sci. doi:10.1098/rspb.2014.0370
Gido KB, Propst DL, Olden JD, Bestgen KR (2013) Multidecadal responses of native and introduced fishes to natural and altered flow regimes in the American Southwest. Can J Fish Aquat Sci 70:554–564
Gilpin ME, Soulé ME (1986) Minimum viable populations: processes of species extinction. In: Soulé ME (ed) Conservation biology: the science of scarcity and diversity. Sinauer Associates Inc, Sunderland, pp 19–34
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Harvey BC, White JL, Nakamoto RJ (2004) An emergent multiple predator effect may enhance biotic resistance in a stream fish assemblage. Ecology 85:127–133
Hernandez-Martich JD, Smith MH (1997) Downstream gene flow and genetic structure of Gambusia holbrooki (eastern mosquitofish) populations. Heredity 79:295–301
Higgins K, Lynch M (2001) Metapopulation extinction caused by mutation accumulation. PNAS 98:2928–2933
Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216
Hillis DM, Mable BK, Larson A, Davis SK, Zimmer EA (1996) Nucleic acids IV: Sequencing and cloning. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn. Sinauer Associates, Sunderland
Hollingsworth PR, Near TJ (2009) Temporal patterns of diversification and microendemism in eastern highland endemic barcheek darters (Percidae: Etheostomatinae). Evolution 63:228–243
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Hughes JM, Schmidt DJ, Finn DS (2009) Genes in streams: using DNA to understand the movement of freshwater fauna and their riverine habitat. Bioscience 59:573–583
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Huxel GR, Hastings A (1999) Habitat loss, fragmentation, and restoration. Restor Ecol 7:309–315
Kindlmann P, Burel F (2008) Connectivity measures: a review. Landscape Ecol 23:879–890
Lewis CA, Lester NP, Bradshaw AD, Fitzgibbon JE, Fuller K, Hakanson L, Richards C (1996) Considerations of scale in habitat conservation and restoration. Can J Fish Aquat Sci 53:440–445
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
McElroy TC, Kandl KL, Trexler JC (2011) Temporal population genetic structure of eastern mosquito fish in a dynamic aquatic landscape. J Hered 102:678–687
McGlashan DJ, Hughes JM, Bunn SE (2001) Within-drainage population genetic structure of the freshwater fish Pseudomugil signifer (Pseudomugilidae) in northern Australia. Can J Fish Aquat Sci 58:1842–1852
Meffe GK, Vrijenhoek RC (1988) Conservation genetics in the management of desert fishes. Conserv Biol 2:157–169
Minckley WL, DeMarais BD (2000) Taxonomy of chubs (Teleostei, Cyprinidae, Genus Gila) in the American Southwest with comments on conservation. Copeia 2000:251–256
Mock KE, Box JCB, Chong JP, Furnish J, Howard JK (2013) Comparison of population genetic patterns in two widespread freshwater mussels with contrasting life histories in western North America. Mol Ecol 22:6060–6073
Morrissey MB, de Kerckhove DT (2009) The maintenance of genetic variation due to asymmetric gene flow in dendritic metapopulations. Am Nat 174:875–889
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
New Mexico Department of Game and Fish (2006) Colorado River basin chubs (roundtail chub Gila robusta, Gila chub Gila intermedia, headwater chub Gila nigra) recovery plan. NMDGF Conservation Services Division, Santa Fe. http://www.wildlife.state.nm.us/conservation/documents/ChubsRecoveryPlan.pdf
Osborne M, Sharp A, Monzingo J, Propst DL, Turner TF (2012) Genetic analysis suggests high conservation value of peripheral populations of Chihuahua chub (Gila nigrescens). Conserv Genet 13:1317–1328
Paroz Y, Propst D, Stefferud J (2006) Long-term monitoring of fish assemblages in the Gila River drainage, New Mexico. Technical Report. New Mexico Department of Game and Fish, Santa Fe, New Mexico
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Peters MB, Turner TF (2008) Genetic variation of the major histocompatibility complex (MHC class II beta gene) in the threatened Gila trout, Oncorhynchus gilae gilae. Conserv Genet 9:257–270
Pilger TJ, Gido KB, Propst DL (2010) Diet and trophic niche overlap of native and nonnative fishes in the Gila River, USA: implications for native fish conservation. Ecol Freshw Fish 19:300–321
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Propst DL (1999) Threatened and endangered fishes of New Mexico. Technical Report No. 1. New Mexico Department of Game and Fish, Santa Fe, New Mexico
Propst DL, Gido KB, Stefferud JA (2008) Natural flow regimes, nonnative fishes, and native fish persistence in arid-land river systems. Ecol Appl 18:1236–1252
Propst DL, Gido KB, Whitney JE, Gilbert EI, Pilger TJ, Monié AM, Paroz YM, Wick JM, Monzingo JA, Myers DM (2014) Efficacy of mechanically removing nonnative predators from a desert stream. River Res Appl. doi:10.1002/rra.2768
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
R Core Team (2012) R: A language and environment for statistical computing. Vienna, Austria. www.r-project.org
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Richter BD, Braun DP, Mendelson MA, Master LL (1997) Threats to imperiled freshwater fauna. Conserv Biol 11:1081–1093
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Schwemm MR (2006) Genetic variation in the Gila robusta complex (Teleostei: Cyprinidae) in the Lower Colorado River. Dissertation, Arizona State University
Seager R, Ting MF, Held I, Kushnir Y, Lu J, Vecchi G, Huang HP, Harnik N, Leetmaa A, Lau NC, Li CH, Velez J, Naik N (2007) Model projections of an imminent transition to a more arid climate in southwestern North America. Science 316:1181–1184
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Stefferud JA, Gido KB, Propst DL (2011) Spatially variable response of native fish assemblages to discharge, predators and habitat characteristics in an arid-land river. Freshw Biol 56:1403–1416
Strayer DL, Dudgeon D (2010) Freshwater biodiversity conservation: recent progress and future challenges. J N Am Benthol Soc 29:344–358
Tessier N, Bernatchez L (1999) Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of landlocked Atlantic salmon (Salmo salar L.). Mol Ecol 8:169–179
Tibbets CA, Dowling TE (1996) Effects of intrinsic and extrinsic factors on population fragmentation in three species of North American minnows (Teleostei: Cyprinidae). Evolution 50:1280–1292
Trujillo JD, Pilger TJ, Douglas MR, Douglas ME, Turner TF (2012) Microsatellite markers for Longfin Dace, Agosia chrysogaster, a sentinel fish species in imperiled arid-land rivers of Sonoran desert. Conserv Genet Resour 4:927–929
Turner TF, Robison HW (2006) Genetic diversity of the caddo madtom, Noturus taylori, with comments on factors that promote genetic divergence in fishes endemic to the Ouachita highlands. Southwest Nat 51:338–345
U.S. Fish and Wildlife Service (2006) Endangered and threatened wildlife and plants; 12-month finding on a petition to list a distinct population segment of the roundtail chub in the lower Colorado River Basin and to list the headwater chub as endangered or threatened with critical habitat. Fed Reg 71:26007–26017
U.S. Fish and Wildlife Service (2012) Endangered and threatened wildlife and plants; endangered status and designations of critical habitat for spikedace and loach minnow: final rule. Fed Reg 77:10810–10932
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vorosmarty CJ, McIntyre PB, Gessner MO, Dudgeon D, Prusevich A, Green P, Glidden S, Bunn SE, Sullivan CA, Liermann CR, Davies PM (2010) Global threats to human water security and river biodiversity. Nature 467:555–561
Wang JL (2009) A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol Ecol 18:2148–2164
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262
Waples RS, Waples RK (2011) Inbreeding effective population size and parentage analysis without parents. Mol Ecol Resour 11:162–171
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Westerling AL, Hidalgo HG, Cayan DR, Swetnam TW (2006) Warming and earlier spring increase western US forest wildfire activity. Science 313:940–943
Whitlock MC, McCauley DE (1990) Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1724
Whitney JE, Gido KB, Propst DL (2014) Factors associated with the success of native and nonnative species in an unfragmented arid-land riverscape. Can J Fish Aquat Sci 71:1134–1145
Whitney JE, Gido KB, Pilger TJ, Propst DL, Turner TF (2015) Metapopulation analysis indicates native and nonnative-fishes respond differently to effects of wildfire on desert streams. Ecol Freshw Fish. doi:10.1111/eff.12217
Wofford JEB, Gresswell RE, Banks MA (2005) Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecol Appl 15:628–637
Acknowledgments
Versions of this manuscript benefitted from comments by N. Franssen and 2 anonymous reviewers. Field and laboratory assistance was provided by T. Diver, N. Franssen, E. Gilbert, E. Martin, A. Monie, J. Monzingo, M. Osborne, J. Perkin, M. Troia, J. Trujillo, and J. Wick. We thank M. Cooper of The Nature Conservancy and the New Mexico Department of Game and Fish (NMDGF) Heart Bar Wildlife Refuge for providing lodging. This research was funded in part by NMDGF Share-with-Wildlife Program and the U.S. Bureau of Reclamation WaterSMART Program (Desert Landscape Conservation Cooperative grant). Tissues were collected under NMDGF permit 3015 (TFT) and followed Institutional Animal Care and Use Committee (IACUC) protocol 10-100492-MCC. We acknowledge logistical help from the Museum of Southwestern Biology Division of Fishes and technical support from the University of New Mexico Molecular Biology Facility, which is supported by NIH Grant P20GM103452.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pilger, T.J., Gido, K.B., Propst, D.L. et al. Comparative conservation genetics of protected endemic fishes in an arid-land riverscape. Conserv Genet 16, 875–888 (2015). https://doi.org/10.1007/s10592-015-0707-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-015-0707-3