Abstract
An endangered population of African wild dogs (Lycaon pictus) disappeared from the Serengeti-Mara area in 1991. The reasons for the extinction are not well understood, but disease was implicated in the disappearance. In 2001, wild dogs naturally re-established themselves in the region. We conducted genetic profiling on samples collected prior and subsequent to this event, as well as samples from three geographically close populations, to determine the potential source of colonisers. Contrary to expectations, we found no evidence of re-colonisation from these nearby wild dog populations. Rather, our analyses suggest that the re-established animals are primarily derived from the same genetic population as the pre-extinction animals, indicating that wild dogs are likely to have persisted in the Serengeti-Mara after 1991. We also detected some migrants that could be derived from genetically distinct populations outside the recovery area. Overall, we did not detect a decline in genetic diversity at either neutral microsatellites or major histocompatibility complex loci, indicating that the supposed disappearance of wild dogs in the Serengeti-Mara did not substantially impact genetic variation of the population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With most of their populations in global decline, there seems little to celebrate in endangered species conservation. One recently heralded exception is the re-establishment of endangered African wild dogs (Lycaon pictus, hereafter, wild dogs) in the Serengeti-Mara area following their apparent extinction in 1991 (Woodroffe 2001). This extinction in the Serengeti-Mara was a high-profile event that spawned a heated debate about its causes, involving more than 20 articles across 9 years (reviewed in Woodroffe 2001). Specifically, concern that, rather than natural causes, human handling of wild dogs in the Serengeti-Mara caused latent rabies to develop and spread (Burrows et al. 1994) led to a ban on animal handling and impeded vaccination programs (Woodroffe 2001). In 2001, wild dogs naturally re-established in the Serengeti-Mara area (Fyumagwa and Wiik 2001). Given the controversial history of this population, there is considerable interest about the origin of these individuals. Since wild dogs are a highly mobile species capable of moving up to 250 km to establish new packs (Fuller et al. 1992), there are a number of potential source populations of the “new” Serengeti-Mara wild dogs. In Eastern Africa, wild dog populations are no longer resident in Uganda, Rwanda or Burundi but extant populations are known in Tanzania and Kenya, as well as further north in Ethiopia and Sudan (Fig. 1a). However, most extant populations are separated from the Serengeti-Mara area not only by distances greater than the recorded dispersal capabilities of the species (>250 km) but also by predominantly uninhabitable and/or human dominated landscape that is associated with high mortality (Woodroffe et al. 2007). Thus, re-colonisation of the Serengeti-Mara area is most likely from the most proximate populations in Tanzania or Kenya.
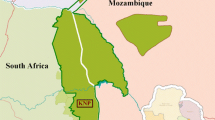
a Map of sampling locations and sample sizes. Green = Laikipia (LAI), blue = Serengeti-Mara (S-M), yellow = Masai Steppe (MST), red = Selous (SEL). Grey circles represent populations that were not sampled in the area surrounding the Serengeti-Mara. b Approximate area of Serengeti-Mara pre-extinction (dark blue) and re-established (light blue) monitoring areas, and Serengeti-Mara ecosystem according to http://www.serengetidata.org/. (Color figure online)
Wild dog monitoring in the Serengeti-Mara area (Fig. 1b) was formerly (pre-extinction) concentrated in two regions: (1) the “Serengeti plains” in the south from 1964 (Burrows et al. 1994); and (2) the “Mara” just outside of the Masai Mara Nature Reserve in the north from 1987 (Scott 1991). Between 1986 and 1991, 15 packs were observed in this pre-extinction monitoring area (Woodroffe 2001). By December 1991, all of the packs previously observed were recorded as absent (Woodroffe 2001) and the entire Serengeti-Mara wild dog population was subsequently reported and widely assumed to be, extinct (Creel and Creel 2002; Daszak et al. 2000; Woodroffe and Ginsberg 1999; but see Ginsberg et al. 1995; Burrows et al. 1994). Outside of the monitoring area, wild dog presence has not been documented systematically, with reports often limited to anecdotal sightings by residents or tourists (Woodroffe 2001). Therefore, high quality data are not available on the location, population size, or demographic trends of wild dogs outside the Serengeti-Mara monitoring area (Burrows 1995). Nonetheless, observations of non resident wild dogs entering the monitoring area between 1965 and 1991 was taken as evidence for the existence of a breeding population “elsewhere” (Burrows et al. 1994; Woodroffe 2001), although not necessarily immediately adjacent to the Serengeti-Mara monitoring area due to the potentially high dispersal abilities of this species (Woodroffe 2001). Limited monitoring conducted in the Serengeti-Mara ecosystem between 1991 and 1998 reported only vagrant and single-sex dispersing groups (Woodroffe 2001). However, following sightings of multiple groups of wild dogs in 1998, systematic monitoring was re-initiated and the Serengeti-Mara population was officially deemed re-established in 2001 when the first denning was reported in the Ngorongoro Conservation Area (Fyumagwa and Wiik 2001). The re-established population has subsequently grown rapidly. Although the home ranges of re-established and pre-extinction packs overlap, the current monitoring area does not include the Serengeti National Park (SNP; Fig. 1b) because, as of Feb 2010, wild dogs have not re-established resident packs there (E. Masenga Pers comm.).
Rarely are samples available prior to extinction and following natural re-colonisation of an endangered species. However, through long-term research programmes, we obtained samples from individuals residing in the Serengeti-Mara area before (pre-extinction) and after the assumed extinction (re-established). These samples, and representatives from three nearby wild dog populations in Eastern Africa (Selous, Masai-Steppe and Laikipia), were genotyped for variation at 10 microsatellite loci to assess the source of the re-established Serengeti-Mara wild dogs. We also assessed whether the re-established Serengeti-Mara population exhibited reduced levels of genetic diversity at both neutral microsatellite loci and at the major histocompatibility complex (MHC), as predicted if founder effects occurred due to re-colonisation by a limited number of wild dogs from outside the Serengeti-Mara area or with a bottleneck associated with a large reduction in the number of animals within the region.
We demonstrate that, despite the observed disappearance of wild dogs in the monitoring area, the declaration of extinction may have been premature, as genetic evidence indicates that the majority of the re-established animals are derived from the Serengeti-Mara population. Encouragingly, we also show that there has not been a loss of genetic diversity in the Serengeti-Mara population.
Materials and methods
Sampling and DNA extraction
We obtained samples from wild dogs residing in the Serengeti-Mara area before (n = 20 from ≥6 packs, S. Cleaveland, P. Kat) and after (n = 13 from 4 packs, M. Emmanuel) the assumed local extinction. We use the term Serengeti-Mara area rather than ecosystem, as some of the collected samples and home ranges of packs fall outside of the official boundaries of the Serengeti-Mara ecosystem, which has been specifically defined by the extent of the wildebeest migration (see Fig. 1b). We also obtained samples from three other wild dog populations in eastern Africa: Selous in southern Tanzania (n = 22 from 8 packs, S. Creel); Masai Steppe in northern Tanzania (n = 32 from 3 packs, A. Visée); and Laikipia in northern Kenya (n = 65 from ≥9 packs, R. Woodroffe; Fig. 1a, b), which are the three geographically closest extant populations for which samples could be obtained. We could not obtain samples from all extant wild dog populations in Kenya and Tanzania because of the absence of monitoring programmes in some areas as well as the general rarity of wild dogs where they still persist (see Fig. 1a). Nonetheless, we did have samples from the closest potential source population, the Masai Steppe. All DNA isolated in this study was originally derived from blood, sera or tissue, and was extracted using a phenol chloroform-based method as in Girman et al. (2001)or using DNeasy tissue and blood extraction kits (Qiagen Inc, Crawley, UK), according to the manufacturer’s instructions.
Microsatellite and MHC genotyping
DNA samples were genotyped at 10 previously published microsatellite loci located on different chromosomes: PEZ08, PEZ12, PEZ15 (J. Halverson in Neff et al. 1999); FHC2010, FHC2054, FHC2611, FHC2658, FHC2785, FHC3399, FHC3965 (Breen et al. 2001; Guyon et al. 2003; Neff et al. 1999). Full details of microsatellite primers are provided in Supplementary Material 1. The forward primer of each pair was labelled with ABI fluorescent dyes: NED (yellow), 6-FAM (blue) or HEX (green), with a ROX 500 size standard. Samples and negative controls were amplified by multiplex PCR using a Qiagen Multiplex PCR mix. We followed default reagent concentrations recommended by the manufacturer, except in cases of DNA derived from serum, where 0.4 μl of 10 mM Bovine Serum Albumin (Promega) was added per PCR reaction. PCR was performed on PTC-200 (MJ Research) thermocyclers with the following touchdown protocol: 15 min at 95°C; 12 touchdown cycles of 94°C for 30 s; 1 min at 30 s annealing, starting at 60°C and reducing at 0.5°C per cycle; and 72°C for 1 min. This was followed by 33 cycles of 89°C for 30 s, 55°C for 1 min, and 72°C for 1 min. A final extension of 60°C for 30 min was performed. Samples were run on an ABI 3730 (by The Sequencing Service, University of Dundee) and analysed using GENEMAPPER 4.7 (Applied Biosystems). Low concentration or poorly amplifying DNA samples were amplified and genotyped three times. Samples with missing data for more than three loci were excluded from analyses. For each population and locus, we tested for deviations from Hardy–Weinberg equilibrium using GENALEX6 (Peakall and Smouse 2006), assessed for significance after Bonferroni correction for multiple tests.
In order to assess whether the re-established populations had suffered from a loss of potentially adaptive genetic diversity, we also compared patterns of diversity at the Major Histocompatibility Complex (MHC). Sequence-based typing was conducted according to the methods outlined in Marsden et al. (2009) and Kennedy et al. (2002). In brief, we amplified exon 2 of the MHC class II DLA-DRB1 locus (hereafter referred to as DRB) by PCR. PCR products were then cleaned using ExoSAP-it (USB), and sequenced directly on an ABI 3730. To determine the alleles present in each individual, DRB sequence data were analysed using Match Tools and Match Tools Navigator (Applied Biosystems). This approach relies on an allele library built from previously identified alleles, which is used to identify the alleles of homozygotes and to predict the most likely allelic combinations present in a heterozygous sequence. We had ten heterozygous individuals that did not match any pair of known alleles, thus indicating the presence of new alleles. Therefore, we cloned DRB PCR products from five animals using the TOPO TA cloning system and One Shot Competent cells (Invitrogen), and sequenced eight clones from each of the five animals. Based on this methodology, we identified three new alleles in this study, which resolved all ten heterozygous sequences. These new alleles were submitted to the DLA nomenclature committee (L.J. Kennedy) to be assigned official names (DRB1*90205, DRB1*90403, DRB1*91101) and have been submitted to GENBANK (accession numbers: JQ085961, JQ085962, JQ085960).
Microsatellite clustering analyses
A neighbour-joining tree based on Nei’s allele-sharing distance was reconstructed in POPULATIONS v 1.2.30 (Langella 1999). Bayesian clustering analysis was conducted using STRUCTURE 2.3.1 (Pritchard et al. 2000), assuming no prior population or location information, with correlated allele frequencies and admixture. We used 100,000 burn-in cycles and 500,000 Markov Chain Monte Carlo (MCMC) runs for K = 1–10, with ten replicates per K value. The most likely number of clusters (K) that best fit the data was selected based on the ∆K statistic (Evanno et al. 2005), calculated through the programme STRUCTURE HARVESTER v0.5 (Earl 2009). The ∆K statistic assesses the rate of change in the log probability of the data between successive K values (Evanno et al. 2005), with the maximal ∆K value taken to indicate the number of clusters at the highest level of structure. However, the presence of secondary peaks is indicative of finer scale structuring; therefore Evanno et al. (2005) suggests running STRUCTURE on both the complete data set, as well as the data subgroups identified by the tallest peak of ∆K, to elucidate finer scale structuring.
Assignment tests
We conducted posterior probability assignment tests on the re-established Serengeti-Mara samples in STRUCTURE 2.3.1. We ran STRUCTURE at K = 4, assuming correlated allele frequencies, admixture, a migration rate of 0.01 (Pritchard et al. 2000), 100,000 burn-in cycles and 500,000 MCMC runs. To consider unsampled populations, we also used Geneclass 2 (Piry et al. 2004), with the Rannala and Mountain criterion (Rannala and Mountain 1997) and Paetkau resampling algorithm (Paetkau et al. 2004), assessed over 10,000 simulations.
Genetic differentiation and diversity
Pairwise population differentiation values were based on the estimator Dest (Jost 2008), calculated with 1000 bootstraps in the programme SMOGD (Crawford 2009). This distance measure was chosen in preference to other frequently used estimators such as FST and GST, because they have been shown not to increase linearly with increasing differentiation (Jost 2008). For both microsatellite and MHC loci we calculated observed (H o ) and expected (H e ) heterozygosity in GENALEX (Peakall and Smouse 2006) and the number of alleles (A) and allelic richness standardized for sample sizes (R s ) in FSTAT 2.9.3 (Goudet 1995).
Results
Clustering analyses
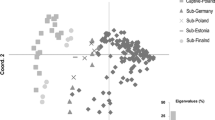
An allele sharing tree indicated that the wild dog samples clustered into four groups corresponding to sampling location: (1) Laikipia; (2) Selous; (3) Masai-Steppe; and (4) pre-extinction and re-established Serengeti-Mara (Fig. 2). Similarly, STRUCTURE analyses were most consistent with four clusters within the data set (Fig. 3a). The ∆K statistic indicated the strongest signal of population subdivision to be K = 2, where Laikipia was distinct from all other samples (Fig. 3a). However, there was a strong secondary peak at K = 4 (Fig. 3a), indicative of finer scale population structuring (Evanno et al. 2005). Therefore, as recommended by Evanno et al. (2005) we re-ran STRUCTURE on each of the clusters indicated at the strongest level of subdivision (K = 2; Fig. 3a): (1) Selous, Serengeti-Mara and Masai-Steppe and (2) Laikipia. For the former, the results showed a clear single peak at K = 3, with the pre-extinction and re-established Serengeti-Mara samples forming one cluster, the Selous samples a second cluster, and Masai Steppe samples a third cluster (Fig. 3b). Independent analysis of Laikipia (cluster 2) detected further subdivision within this population (n = 65 from ≥9 packs), with individuals clustering into four groups of related packs (Fig. 3c). The high level of distinction of Laikipia relative to the other wild dog populations at K = 2 indicates this to be a very divergent population.
Population subdivision based on neighbour-joining tree of allele-sharing distance across 10 microsatellite loci. Colours and abbreviations correspond to Fig. 1 with the exception of the re-established Serengeti-Mara samples which are coloured brown to distinguish them from the pre-extinction Serengeti-Mara samples (blue). Asterisks depict pre-extinction Serengeti-Mara samples from the northern Mara area. Re-established Serengeti samples (brown) clearly cluster with pre-extinction Serengeti samples rather than with the three other populations. (Color figure online)
Hierarchical population structure based on STRUCTURE analyses of 10 microsatellite loci. The columns in the bar charts represent individuals, with the proportion of an individual’s genotype assigned to each cluster (K) denoted by different colours. Shown are the most likely number of clusters (K) according to the ∆K statistic (line graphs) at the uppermost (a) and finer scale (b, c) levels of structuring. Pre-ext = Pre-extinction Serengeti-Mara; Re-est = Re-established Serengeti-Mara. Putative migrants are indicated with asterisks based on individual genotype assignments of <70% to their sampled population (b). For analysis within Laikipia (c), individuals were sorted by pack membership depicted with numbers (1–14). Individuals within unknown pack membership were grouped as pack 7. (Color figure online)
Despite our findings of strong population structure, the presence of potential migrants was indicated by clustering analyses in Selous, Masai Steppe, pre-extinction Serengeti-Mara and Laikipia by mis-assignments in the allele sharing tree and STRUCTURE. Specifically, in the allele sharing tree (Fig. 2), sample WDM22 from Laikipia and sample RKW328 from the pre-extinction Serengeti-Mara clustered with the Selous rather than their respective populations, and sample RKW12165 from Selous clustered with Masai Steppe. In STRUCTURE (Fig. 3a), three samples exhibited low (<70%) ancestry values for their respective populations; specifically sample MK263 from Masai Steppe and RKW328 from the pre-extinction Serengeti-Mara. Both these individuals had high ancestry proportions to Selous. Moreover, RKW 12156 from Selous appeared to be admixed.
Assignment tests
Based on microsatellite loci, posterior probability assignment tests implemented in STRUCTURE showed that individuals from the re-established Serengeti-Mara population had on average a 95% probability of assignment to the same population as the pre-extinction animals (range 85–97%), compared with 1.4–2.4% probability of assignment to any of the other putative source populations. The more conservative assignment test that allows for unsampled populations, implemented in GENECLASS2, assigned the majority (9/13) of re-established Serengeti-Mara animals to the same population as the pre-extinction individuals. However, it is noteworthy that this analysis would be unable to detect migration from an unsampled population genetically similar to the pre-extinction Serengeti-Mara population. The remaining four individuals were assigned to unsampled populations, indicating that they could be migrants from a population outside of the Serengeti-Mara monitoring area that we did not sample. It is also possible that our sampling was not extensive enough to exhaustively sample the allelic diversity present in the pre-extinction samples and so we cannot exclude the possibility that the putative migrants were actually from the same gene pool as the pre-extinction population. Unfortunately, we used all available pre-extinction samples so the likelihood of this possibility cannot be assessed at this time.
Genetic differentiation and diversity
We observed no significant decline in H o , H e , or R s at microsatellite loci in the re-established population relative to the pre-extinction population (Table 1: paired T test; H o , t = 0.72, P = 0.486; H e , t = 1.90, P = 0.09; R s , t = 1.70, P = 0.123). Overall, 79% of microsatellite alleles detected in the re-established population were also detected in the pre-extinction population. In general, levels of genetic diversity were comparable to the three other wild dog populations (Table 1) as well as other wolf-like canid populations (Aspi et al. 2006; Aspi et al. 2009; Randall et al. 2010). No significant deviations from Hardy–Weinberg equilibrium at any microsatellite locus were observed (GENALEX6 Peakall and Smouse 2006). Pairwise genetic differentiation comparisons (Dest) based on microsatellite loci showed the re-established Serengeti-Mara to be most similar to the pre-extinction Serengeti-Mara (Dest = 0.14; other populations, 0.25–0.47; Table 2).
Across populations, we amplified nine different MHC class II DRB alleles (Table 3), and found no evidence of pseudogenes (stop codons or frameshift mutations), as well as a maximum of two alleles amplified per individual, suggesting that a single functional gene was assayed. As with the microsatellite data, MHC diversity in the re-established populations was comparable to the three other wild dog populations and we found no evidence of a decline in MHC diversity in the re-established relative to the pre-extinction Serengeti-Mara population (Table 1). In fact, H o , H e and R s were all higher in the re-established population. However, there was a shift in allelic composition and frequency between pre-extinction and re-established samples (Table 3). As such, the pre-extinction and re-established populations shared just 22% of MHC alleles (2/9; Table 3). For example, allele DRB1*90601 was found at high frequency (33%) in the pre-extinction population but was absent in the re-established population. Similarly, allele DRB1*90203, which was common (43%) in the re-established population, was not found in the pre-extinction population. Nonetheless it is noteworthy that allele DRB1*90204, which is unique to the pre-extinction Serengeti-Mara, was also found in the re-established population in three of the four sampled packs. In comparison to other populations, the re-established samples shared the most MHC alleles with the Selous population (30%, 4/13; Table 3). Overall, however, the re-established Serengeti-Mara was highly differentiated from all populations at the MHC (Dest = 0.44–0.75), which contrasts with the pattern at microsatellite loci (Table 2), as might be expected for genes that could be subject to both balancing and directional selection (reviewed in Bernatchez and Landry 2003).
Discussion
The Serengeti-Mara wild dog population disappeared in 1991. However, in 2001, a denning pack was observed, indicating that a breeding population had re-established (Fyumagwa and Wiik 2001). We conducted genetic profiling on animals from both the pre-extinction and re-established Serengeti-Mara populations, as well as animals from nearby populations in East Africa, to distinguish between three possible scenarios: (1) the Serengeti-Mara wild dog population became extinct in 1991 and the re-established animals migrated from another genetically distinct wild dog population; (2) some of the Serengeti-Mara wild dog population persisted undetected post 1991, and the re-established population is derived from the descendants of these animals; (3) the Serengeti-Mara wild dogs represent a mixture of migrants and descendants of the pre-extinction population.
Contrary to expectations if there had been an extinction event, both Bayesian and allele-sharing clustering analyses showed that re-established Serengeti-Mara wild dogs grouped with pre-extinction individuals rather than other putative source populations (Figs. 2, 3). Furthermore, assignment tests indicated that the majority of re-established Serengeti-Mara wild dogs were derived from the same genetic population as the pre-extinction Serengeti-Mara population. Lastly, genetic differentiation based on neutral markers showed the re-established Serengeti-Mara to be most similar to the pre-extinction Serengeti-Mara. Consequently, these findings strongly imply that although wild dogs were no longer observed to be resident in the well-monitored areas of the Serengeti-Mara after 1991, they persisted undetected there, or in unmonitored areas nearby. It has been widely assumed that all former residents (15 packs) in the pre-extinction monitoring area died, but the actual fates of most are unknown (Ginsberg et al. 1995; Woodroffe 2001). Passive monitoring by scientists, tour guides, and indigenous communities suggests an absence of breeding packs from the pre-extinction monitoring area until at least 1998. However, these monitoring efforts may not have been sufficient to detect a population present at low density, especially given the inaccessible terrain and dense vegetation cover in parts of the monitoring area. Furthermore, surveys for wild dogs were limited and sightings largely anecdotal in localities just outside of the monitoring areas, such as in the Loliondo Game Control Area (LGCA), where much of the re-established population currently resides. Indeed, Burrows et al. (1994) suggested that evidence of non-resident wild dogs continuing to enter the pre-extinction monitoring area after 1991 (4.5 ± 3.5 dogs/year) was indicative of breeding packs nearby the pre-extinction monitoring area (but see Woodroffe 2001). The presence of resident wild dogs near to the pre-monitoring area after 1991 is indicated by observations of wild dogs denning in the Loita Hills in 1995 (Martyn 1995), which is approximately 30 km east of the northern part of the pre-extinction monitoring area. By contrast, wild dogs are not currently found to the west of the Serengeti-Mara near Lake Victoria in Tanzania or in Uganda (IUCN/SSC 2008) and colonisation seems less likely from the North, given the high genetic differentiation between Laikipia and Serengeti-Mara.
Although our results strongly imply that most of the re-colonisers were derived from the same genetic population as the pre-extinction population. The assignment of four individuals to unsampled populations by GENECLASS2 suggest that some of the re-established Serengeti-Mara could be migrants from elsewhere (although as explained above we cannot exclude the possibility that the putative migrants were actually from the pre-extinction population). As detailed in the results, the presence of migrants was also indicated in other wild dog populations by mis-assignments in the allele sharing tree (Fig. 2) and STRUCTURE (Fig. 3). In one of these cases, WDM 22 from Laikipia, field information also suggested this individual to be a migrant. However, for all other examples, field information was either unavailable or did not suggest migrant ancestry. Given the large distance between pre-extinction Serengeti-Mara, Laikipia and Selous (>600 km), these migrants most likely came from an unsampled more proximate population.
Rarely are genetic samples before and after a local demographic decline in an endangered species available and their existence in this study highlights the importance of continuous long-term field projects such as those in the Serengeti-Mara (Thirgood et al. 2007). Although our sample size is small, we found no evidence of a loss of genetic diversity at neutral microsatellite loci or genes that should be subject to selection (MHC-DRB), suggesting that the apparent disappearance of wild dogs in the Serengeti-Mara did not impact genetic variation of the larger population (Table 1). In addition to rapid population expansion (vonHoldt et al. 2010) and inbreeding avoidance (Von Holdt et al. 2008), migration from other genetically distinct populations (as suggested by GENECLASS) may have helped to maintain diversity despite genetic drift.
We did detect a shift in allele composition and increased heterozygosity at the MHC in the re-established population that was not mirrored at neutral markers. Due to the functional role of the MHC in immunity, these changes could represent adaptive differences between the pre-extinction and re-established study populations resulting from changes in selective pressures. Indeed, allelic composition and heterozygosity were more consistent at neutral loci (52% of alleles shared pre- and post-extinction, Ho–4% decrease) than at DRB (22% of alleles shared, Ho–26% increase). Unfortunately, insufficient data exist to determine whether these changes are in anyway related to the disease epidemic that was implicated in the population crash (reviewed in Woodroffe 2001).
We found that three of the six MHC alleles of the re-established Serengeti-Mara were unique to individual packs and similar proportions were found for the other populations where we had pack information for all individuals: Selous, 5/11; Masai Steppe, 2/5. This apparent spatial structuring of MHC alleles could be the result of sampling effects associated with the social structure of this species, since wild dog packs consist of close relatives (Girman et al. 2001). Such sampling effects may explain some of the changes in allele composition between the pre-extinction and re-established samples. A small number of breeders (Luikart and Cornuet 1999), or bottleneck effects (Cornuet and Luikart 1996) could also have contributed to changes in the frequency of MHC alleles. Based on the neutral microsatellite markers, results from GENECLASS2 indicated the presence of putative immigrants in the re-established population, and these individuals could have introduced new MHC alleles from an unsampled population. This appears to be the case for one of the putative migrants, SFMUT, who was the only individual with allele, DRB1*91101. The three remaining putative migrants did not possess any unique alleles. However, they were migrants from packs where other wild dogs, and thus potentially their offspring with whom they would share unique alleles, had been sampled. Overall, the re-established population share a large fraction of MHC alleles with the other wild dog populations (Table 3). A high degree of allele sharing is not unexpected because the MHC is under balancing selection and alleles are often shared between populations (or species) for long periods of time (trans-specific polymorphism; Klein 1987). Thus, only the neutral data are useful for inferring genetic connectivity among populations.
Our study is informative about the origin of the ‘re-colonisers’. However, genetic data alone cannot provide information on the puzzling cause of the disappearance of so many packs in the Serengeti-Mara monitoring area, nor on reasons for their subsequent recovery. More extensive monitoring would have been required to evaluate the causes of the wild dog disappearance (Woodroffe 2001). Nonetheless, it is interesting to note that, to date, no packs have re-established in the Serengeti National Park where much of the pre-extinction monitoring was focused (Fig. 1b), despite observations of sporadic incursions of individuals into this area (M. Emmanuel Pers comm.). The reason for the absence of wild dogs from the Serengeti National Park remains to be determined but they are known to avoid lions (Mills and Gorman 1997), which have increased inside the park since 1991 (Packer et al. 2005).
In conclusion, our data suggest that the re-established Serengeti-Mara population represents a mixture of descendants of the pre-extinction Serengeti-Mara population as well as a smaller number of migrants from genetically more distant populations (scenario 3). Furthermore, we have shown that the supposed disappearance of wild dogs in the Serengeti-Mara did not strongly impact genetic diversity of the local population.
References
Aspi J, Roininen E, Ruokonen M, Kojola I, Vila C (2006) Genetic diversity, population structure, effective population size and demographic history of the Finnish wolf population. Mol Ecol 15:1561–1576
Aspi J, Roininen E, Kiiskilä J, Ruokonen M, Kojola I, Bljudnik L, Danilov P, Heikkinen S, Pulliainen E (2009) Genetic structure of the northwestern Russian wolf populations and gene flow between Russia and Finland. Conserv Genet 10:815–826
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J Evol Biol 16:363–377
Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, Holmes NG, Chéron A, Suter N, Vignaux F, Bristow AE, Priat C, McCann E, André C, Boundy S, Gitsham P, Thomas R, Bridge WL, Spriggs HF, Ryder EJ, Curson A, Sampson J, Ostrander EA, Binns MM, Galibert F (2001) Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res 11:1784–1795
Burrows R (1995) Demographic changes and social consequences in wild dogs, 1964–1992. In: Sinclair ARE, Arcese P (eds) Serengeti II: dynamics, management and conservation of an ecosystem. University of Chicago Press, Chicago, pp 400–420
Burrows R, Hofer H, East ML (1994) Demography, extinction and intervention in a small population: the case of the serengeti wild dogs. Proc R Soc Lond B 256:281–292
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crawford NG (2009) SMOGD: software for the measurement of genetic diversity. Mol Ecol Res 10:556–557
Creel S, Creel NM (2002) The African wild dog: behavior, ecology and conservation. Princeton University Press, Princeton, NJ
Daszak P, Cunningham AA, Hyatt AD (2000) Emerging infectious diseases of wildlife -threats to biodiversity and human health. Science 287:443–449
Earl DA (2009) Structure harvester v.0.5. Available from http://taylor0.biology.ucla.edu/struct_harvest/
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fuller TK, Mills MGL, Borner M, Laurenson K, Kat PW (1992) Long distance dispersal by African wild dogs in east and south Africa. J. Afr Zool 106:535–537
Fyumagwa D, Wiik H (2001) TAWRI Wildlife veterinary programme Annual Report
Ginsberg J, Mace G, Albon S (1995) Local extinction in a small and declining population: wild dogs in the serengeti. Proc R Soc Lond B 262:221–228
Girman DJ, Vila C, Geffen E, Creel S, Mills MGL, McNutt JW, Ginsberg J, Kat PW, Mamiya KH, Wayne RK (2001) Patterns of population subdivision, gene flow and genetic variability in the African wild dog (Lycaon pictus). Mol Ecol 10:1703–1723
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, Parker HG, Quignon P, Lowe JK, Renier C, Gelfenbeyn B, Vignaux F, DeFrance HB, Gloux S, Mahairas GG, André C, Galibert F, Ostrander EA (2003) A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci USA 100:5296–5301
IUCN/SSC (2008) Regional conservation strategy for the cheetah and wild dog in eastern Africa. IUCN, Gland
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kennedy LJ, Barnes A, Happ GM, Quinnel RJ, Courtenay O, Carter SD, Ollier WER, Thomson W (2002) Evidence for extensive DLA polymorphism in different dog populations. Tissue Antigens 60:43–52
Klein J (1987) Origin of the major histocompatibility complex polymorphism: the trans-species hypothesis. Hum Immunol 19:155–162
Langella O (1999) POPULATIONS v.1.2.30. 1.2.30
Luikart G, Cornuet J-M (1999) Estimating the effective number of breeders from heterozygote excess in progeny. Genetics 151:1211–1216
Marsden CD, Mable BK, Woodroffe R, Rasmussen GSA, Cleaveland S, McNutt JW, Emmanuel M, Thomas R, Kennedy LJ (2009) Highly endangered African wild dogs (Lycaon pictus) lack variation at the major histocompatibility complex. J Hered 100:S54–S65
Martyn N (1995) The Naikara wild dogs. East Afr Nat His Soc Bull 24
Mills MGL, Gorman ML (1997) Factors affecting the density and distribution of wild dogs in the Kruger National Park. Conserv Bio 11:1397–1406
Neff MW, Broman KW, Mellersh CS, Ray K, Acland GM, Aguirre GD, Ziegle JS, Ostrander EA, Rine J (1999) A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics 151:803–820
Packer C, Hilborn R, Mosser A, Kissui B, Borner M, Hopcraft G, Wilmshurst J, Mduma S, Sinclair ARE (2005) Ecological change, group territoriality, and population dynamics in Serengeti lions. Science 307:390–393
Paetkau D, Slade R, Burden M, Estoup A (2004) Direct, real-time estimation of migration rate using assignment methods: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Res 6:288–295
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: A software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Randall D, Pollinger J, Argaw K, Macdonald D, Wayne R (2010) Fine-scale genetic structure in Ethiopian wolves imposed by sociality, migration, and population bottlenecks. Conserv Gen 11:89–101
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Scott J (1991) Painted wolves: wild dogs of the Serengeti-Mara. Hamish Hamilton, London
Thirgood SJ, Mduma SAR, Keyyu JD, Laurenson MK (2007) The value of long-term research: conservation science and practice in Tanzania. Conserv Bio 21:576–579
Von Holdt BM, Stahler DR, Smith DW, Earl DA, Pollinger JP, Wayne RK (2008) The genealogy and genetic viability of reintroduced yellowstone grey wolves. Mol Ecol 17:252–274
vonHoldt BM, Stahler DR, Bangs EE, Smith DW, Jimenez MD, Mack CM, Niemeyer CC, Pollinger JP, Wayne RK (2010) A novel assessment of population structure and gene flow in grey wolf populations of the Northern Rocky Mountains of the United States
Woodroffe R (2001) Assessing the risks of intervention: immobilization, radio-collaring and vaccination of African wild dogs. Oryx 35:234–244
Woodroffe R, Ginsberg JR (1999) Conserving the African wild dog (Lycaon pictus). I. Diagnosing and treating causes of decline. Oryx 33:132–142
Woodroffe R, Davies-Mostert H, Ginsberg J, Graf J, Leigh K, McCreery K, Robbins R, Mills G, Pole A, Rasmussen G, Somers M, Szykman M (2007) Rates and causes of mortality in endangered African wild dogs Lycaon pictus: lessons for management and monitoring. Oryx 41:215–223
Acknowledgement
We thank S. Cleaveland, S. Creel, P. Kat, M. Emmanuel, R. Woodroffe and A. Visée for providing samples and/or field information. We are grateful to L. Kennedy, D. Greenfield and A. Adam for assistance with MHC and microsatellite genotyping and S. Cleaveland, R. Woodroffe and two anonymous reviewers for comments on earlier versions of the manuscript. Funding was provided by a PhD studentship from the Natural Environment Research Council and Royal Zoological Society of Scotland, Edinburgh zoo (NER/S/A/2006/14139).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Marsden, C.D., Wayne, R.K. & Mable, B.K. Inferring the ancestry of African wild dogs that returned to the Serengeti-Mara. Conserv Genet 13, 525–533 (2012). https://doi.org/10.1007/s10592-011-0304-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0304-z