Abstract
Objectives
To develop an in vitro method for rapid evaluation of the capability of a designed single guide RNAs (sgRNAs) to guide Cas9 nucleases to cleave target loci in mammalian cells.
Results
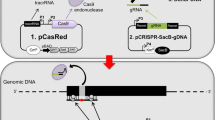
We constructed a Cas9/sgRNA plasmid with two SP6 promoters to simultaneously express Cas9 nuclease and the sgRNA and a negative selection plasmid harbouring a target site of the sgRNA. After co-transforming chemically competent E. coli DH5α cells with the two plasmids, the transformants were plated at a low density on two LB plates: one containing only ampicillin and the other containing both ampicillin and chloramphenicol. The colony-count on the ampicillin + chloramphenicol plate was compared with that on the ampicillin-only plate to calculate the survival percentage. The survival % was negatively correlated with the genome editing efficiency of the sgRNA in mammalian cells evaluated by a T7 endonuclease 1 (T7E1) assay (r ranged from −0.8 to −0.92). This system eliminates the need for cell culture, transfection, FACS sorting, PCR and T7E1 nuclease treatment, and significantly reduces the cost of screening for active sgRNAs, especially in the case of large-scale screening.
Conclusions
We have developed a bacterial-based negative selection system for rapid screening of active sgRNAs in mammalian cells at a very low cost.






Similar content being viewed by others
References
Cong L, Ran FA, Cox D, Lin S, Barretto R et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C et al (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400
Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M et al (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32:1262–1267
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R et al (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT et al (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485
Lee JS, Kallehauge TB, Pedersen LE, Kildegaard HF (2015) Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Sci Rep 5:8572
Luo ML, Leenay RT, Beisel CL (2016) Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng 113:930–943
Mei Y, Wang Y, Chen H, Sun ZS, Ju XD (2016) Recent progress in CRISPR/Cas9 technology. J Genet Genomics 43:63–75
Peters JM, Colavin A, Shi H, Czarny TL, Larson MH et al (2016) A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506
Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK et al (2014) Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24:1020–1027
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA et al (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK et al (2010) ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acid Res 38:W462–W468
Vouillot L, Thelie A, Pollet N (2015) Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5:407–415
Acknowledgements
This work was jointly supported by National Transgenic Major Program (2016ZX08006003-006) and the Natural Science Foundation of Guangdong Province (2016A030313310).
Supporting information
Supplementary Table 1—sgRNA oligos for cloning into pX330-SP6.
Supplementary Table 2—sgRNA oligos for cloning into pX458.
Supplementary Table 3—Primers used for T7E1 assay.
Supplementary Table 4—Statistical analysis of colony formation in bacterial-based negative selection system.
Supplementary Fig. 1—Sequence analysis of cloned PCR products from cells transfected withCas9/single guide RNA (sgRNA) expression plasmids.
Supplementary Fig. 2—Correlation between insertions or deletions (indel) rate reflected by TA-cloning sequencing results and bacterial survival rate.
Supplementary Fig. 3—Transformation results of the bacterial-based negative selection assay.
Supplementary Fig. 4—Leaky expression of EGFP in E. coli DH5α cells under the control of SP6 promoter.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nie, Y., Cong, P., Liu, X. et al. Development of a bacterial-based negative selection system for rapid screening of active single guide RNAs. Biotechnol Lett 39, 351–358 (2017). https://doi.org/10.1007/s10529-016-2259-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2259-0