Abstract
Objectives
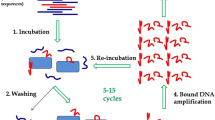
To select aptamers for endotoxin separation from a 75-nucleotide single-stranded DNA random library using systematic evolution of ligands by exponential enrichment.
Results
After 15 rounds of selection, the final pool of aptamers was specific to endotoxin. Structural analysis of aptamers that appeared more than once suggested that one aptamer can form a G-quartet structure. Tests for binding affinity and specificity showed that this aptamer exhibited a high affinity for endotoxin. Using this aptamer, aptamer-magnetic beads were designed to separate endotoxin.
Conclusions
Using these aptamer-magnetic beads, a new method to separate endotoxin was developed to enable specific separation of endotoxin that can be applied to drug and food products.






Similar content being viewed by others
References
Chauveau F, Pestourie C, Tavitian B (2006) Aptamers: selection and scope of applications. Pathol Biol 4:251–258
Hu P, Liu ZS, Tian RY, Ren HL, Wang XX, Lin C, Gong S, Meng XM, Wang GM, Zhou Y, Lu SY (2013) Selection and identification of a DNA aptamer that mimics saxitoxin in antibody binding. J Agric Food Chem 14:3533–3541
Ikeda T, Ikeda K, Suda S, Ueno T (2014) Usefulness of the endotoxin activity assay as a biomarker to assess the severity of endotoxemia in critically ill patient. Innate Immun 8:881–887
Li Y, Lander R, Manger W, Lee A (2004) Determination of lipid profile in meningococcal polysaccharide using reversed-phase liquid chromatography. J Chromatogr B 804:353–358
Missailidis S, Thomaidou D, Borbas KE, Price MR (2005) Selection of aptamers with high affinity and high specificity against C595, an anti-MUC1 IgG3 monoclonal antibody, for antibody targeting. J Immunol Methods 1:45–62
Nutiu R, Li YF (2005) Aptamers with fluorescence-signaling properties. Methods 1:16–25
Petsch D, Anspach FB (2000) Endotoxin removal from protein solutions. J Biotechnol 76:97–119
Satoru N, Noburu I (2011) Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie 93:1231–1238
Tabarzad M, Kazemi B, Vahidi H, Aboofazeli R, Shahhosseini S, Nafissi-Varcheh N (2014) Challenges to design and develop of DNA aptamers for protein targets. I. Optimization of asymmetric PCR for generation of a single stranded DNA library. Iran J Pharm Res 13:133–141
Warren JR (1982) Polymyxin B suppresses the endotoxin inhibition of concanavalin a-mediated erythrocyte agglutination. Infect Immun 2:594–599
Wilson MJ, Haggart CL, Gallagher SP, Walsh D (2001) Removal of tightly bound endotoxin from biological products. J Biotechnol 88:67–75
Yamamoto A, Ochiai M, Kataoka M, Toyoizumi H, Horiuchi Y (2002) Development of a highly sensitive in vitro assay method for biological activity of endotoxin contamination in biological products. Biologicals 2:85–92
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ying, G., Zhu, F., Yi, Y. et al. Selecting DNA aptamers for endotoxin separation. Biotechnol Lett 37, 1601–1605 (2015). https://doi.org/10.1007/s10529-015-1839-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1839-8