Abstract
Musculoskeletal models permit the determination of internal forces acting during dynamic movement, which is clinically useful, but traditional methods may suffer from slowness and a need for extensive input data. Recently, there has been interest in the use of supervised learning to build approximate models for computationally demanding processes, with benefits in speed and flexibility. Here, we use a deep neural network to learn the mapping from movement space to muscle space. Trained on a set of kinematic, kinetic and electromyographic measurements from 156 subjects during gait, the network’s predictions of internal force magnitudes show good concordance with those derived by musculoskeletal modelling. In a separate set of experiments, training on data from the most widely known benchmarks of modelling performance, the international Grand Challenge competitions, generates predictions that better those of the winning submissions in four of the six competitions. Computational speedup facilitates incorporation into a lab-based system permitting real-time estimation of forces, and interrogation of the trained neural networks provides novel insights into population-level relationships between kinematic and kinetic factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human movement is a feat of extraordinary skill, requiring the coordinated action of huge numbers of muscle elements, in the face of changing environmental conditions, in real time. Exactly how the central nervous system (CNS) is able to achieve this task is unclear. In particular, the decoding of the neural representation of abstract task-related goals such as movement into specific muscle activation signals has proven difficult to decipher.
An approximation of this mapping is central to attempts at estimating the magnitudes of internal loads during movement, a matter of established clinical relevance for many patient groups.25 In the field of biomechanics, musculoskeletal models are used to approximate the mapping from kinematic space to force space by applying a framework rooted in the laws of classical mechanics. Experimentally observed kinematics and external forces can be used to formulate the equations of motion, the solution of which yields intersegmental forces and torques. It is in attributing these torques to individual muscles—the load-sharing problem—that it is necessary to specify the motor strategy used by the CNS. This is usually done by minimisation of an objective function representing some putative overarching goal of the motor system, with numerical optimisation techniques used to arrive at an optimal solution.
The use of these techniques has been validated to produce reasonable estimates of joint contact forces and muscular activation patterns for repetitive motions such as gait16,26 but it is difficult to offer a biologically consistent rationale for the choice of any objective function, given our lack of knowledge about the method used by the CNS. Moreover, for complex models in high-dimensional spaces the computational complexity—and thus slowness—of static optimisation makes it a poor fit given the speed with which humans can accomplish complex movements. This is of practical importance as it limits the utility of modelling for real time applications, where the potential for clinical benefit is great.
In addition, as a technique that deals with separate trials independently, traditional inverse dynamic/ static optimisation analysis has no capacity to exploit population-wide relationships between input and output, standing in contrast to models built upon data from multiple trials.
Supervised models of motor learning
Human motor learning involves adaptation to changing conditions – in both the environment and the plant (such as those due to growth, training or disease), and the modulation of muscle activations to account for such changes. For example, upon exposure to a force field, human subjects demonstrate modification of muscular activation patterns to efficiently compensate for the motor disturbance,34 with persistence of learned effects for some time following disengagement from the field. These observations lend weight to the notion of internal models that contain representations of the physics of the external world and of the musculoskeletal system, as put forward by Wolpert et al.41 These models must be updated dynamically to reduce errors such that future movements made on the basis of their predictions are more optimal. This framing of motor control as a problem of supervised learning has its roots in the work of Albus and Marr,2,23 where it was applied specifically to the cerebellum. Later, Kawato et al. proposed hierarchical neural networks as a solution to the control problem and showed that an internal inverse dynamics model could be learned by a robotic controller.15
More recently, supervised learning, manifest as neural networks, has been used to obtain increasingly impressive performances in a number of challenging prediction problems such as object and speech recognition, outperforming traditional modelling devices.11,19 These breakthroughs have been viewed as significant because they represent foundational problems of human intelligence—tasks at which humans naturally excel but which have traditionally proven difficult for machines—much like the motor control problem. In the motor system, hierarchical layering of the type found in neural networks has been proposed as a means of mitigating problems associated with the immense complexity of the control problem,43 although it remains poorly understood.
Application of Supervised Learning to Musculoskeletal Modelling
The replacement of a computationally expensive optimisation process with a fast solution learned under supervision is a well-established use of supervised models. There have been multiple applications of such ‘surrogate’ models both within the domain of biomechanical simulation—with uses in the estimation of implant pressure distribution4 and deformable joint contact8—as well as elsewhere.13 A major advantage of these models is speed; training may be lengthy but as inference involves a relatively simple forward pass through the network, it is computationally inexpensive and thus very quick. The potential for computational speedup is important as the slowness of static optimisation can make real time estimation of internal forces with complex models difficult. The problem of modelling complexity has been surmounted by the use of proxies for internal forces, such as the external knee adduction moment,40 an important biomarker for patients with osteoarthritis (OA) of the knee. As prevalence rates for this disease soar in the western world39 novel preventative approaches have been sought, amongst them real time gait retraining, which aims to slow disease progression through the entraining of a walking technique that mitigates joint loading. However, evidence of a significant disparity between the external adduction moment and true internal forces36 emphasises the need for true force estimation. This has been attempted in real time by explicit limitation of model complexity. Van den Bogert et. al.37 achieved a real time system that employed static optimisation to produce muscle force estimates at a rate in excess of 100 Hz but in order to achieve such speed muscle moment arms were approximated by prescribed polynomials and an iterative optimisation solver was curtailed after a set period of time had elapsed. More recently, EMG-driven models have been run close to real time. One such system was used to provide feedback on kinetic variables to subjects walking on a treadmill, but with a latency of 115 ms,27 far in excess of the maximum 75ms considered optimal for real time biofeedback.14
Further benefits stem from the capacity of machine learning techniques to find latent structure in data. This leads to an important possibility: a reduction in the need for input data in the formulation of predictive models. The extensive ensemble of data required for the operation of physics-based musculoskeletal is an important factor limiting the clinical utilisation of such models.
Finally, because networks are imprinted with the training data, which contribute to their final nodal weights, their use allows a probing of population-level causal relationships between inputs and outputs. The trained network may be viewed as a function approximator where the information encoded in population-level relationships has been transferred to its weights, and may be analysed through interrogation of the network.
In addition to the surrogate models described above, previous attempts to apply neural networks within the field of musculoskeletal modelling have involved mappings between EMG and kinematics,10,33 and EMG to force.20,33 However, datasets have been small and the networks employed necessarily simple and shallow, limiting the generalisability of the results obtained. More recently, deep models combining recurrent and convolutional modules have been applied to time series of biological signals, for example in the prediction of kinematic parameters from EMG data.42 There has also been growing interest in the use of deep learning architectures for motion capture-independent pose estimation.24
Inspired by the recent successes of deep learning for prediction problems, the present study aimed to exploit a large corpus of motion data, applying deep neural networks to compute the mapping from kinematic to muscle space. Two sources of labelled data were used: musculoskeletal modelling predictions obtained by inverse dynamic analysis and static optimisation, and EMG sensor data. Validation was performed using holdout data subsets. Further validation of the models thus produced was performed using in-vivo knee prosthetic data previously used for the ‘Grand Challenge’ competitions to predict in-vivo knee loads.18
Materials and methods
The dataset comprised synchronously captured kinematic (lower limb marker trajectories obtained by optoelectronic capture—Vicon MX system, Vicon Motion Systems Ltd, Oxford, UK), force plate (ground reaction force and centre of pressure—Kistler Instrumente AG, Winterthur, Switzerland) and EMG (Trigno Wireless EMG system, Delsys, USA) data from 156 subjects during multiple trials of level walking.21 Stance phase of both left and right lower limb were represented and treated equivalently. EMG signals were measured for 8 major muscles of each lower limb. The subjects were diverse in body morphology, age, gender and lower limb pathology, with a significant proportion suffering from clinically diagnosed OA of one or both knees (Table 1).
Several prediction problems were attempted:
-
Prediction of medial knee joint reaction force
-
Prediction of forces for major muscle groups of the lower limb
-
Prediction of EMG sensor measurements
Input Tensors for Neural Network Training
Raw data comprising 3-dimensional marker positions for each of 18 lower limb markers (4 on the pelvis and 14 on the stance limb), and force plate-derived 3-dimensional centre of pressure and ground reaction force were extracted from the duration of stance phase for each trial. The data were used to train neural networks taking the data of a single time step of a single trial as input. Thus, where the entirety of the data were utilised, kinematic and kinetic data at each time step were combined into a single vector of length 60, and then concatenated with equivalent vectors from all time steps, t, of a given trial to produce a matrix of dimensions [t, 60]. Many such matrices from all processed gait trials were concatenated to form a single long matrix representing the whole dataset for the neural network, where each row represented a single input.
In the first instance, both kinematic and kinetic data were used for training. Then, models were built using only partial inputs in order to assess the ability of the trained models to produce accurate predictions without exposure to the full ensemble of input data.
Target Tensors for Neural Network Training
Inverse dynamic modelling was used to provide targets for training. Processing of raw motion data was performed using Vicon Nexus® (1.85) and Matlab® (2017a; The MathWorks Inc., Natick, MA, USA). Data filtering was performed in Matlab using a low-pass fourth order Butterworth filter with a cutoff frequency of 10 Hz. Freebody (v2.1),5 an open-source segment-based musculoskeletal model was used for subsequent data processing to determine internal forces. The model’s predictions of tibiofemoral JRF during gait have been validated using data from instrumented prostheses,7 and predicted muscle force waveforms have been shown to demonstrate high levels of concordance with known electromyography envelopes.5,31 Implementation involved the determination of coordinates of internal points in a subject-specific frame of reference. This was achieved by scaling using the measurements of a gender and height-matched subject, chosen from a morphologically diverse cohort of eight subjects, for whom three-dimensional position data of internal points had been obtained using magnetic resonance imaging.7 Processed data were then taken as input by a Matlab® implementation of interior points optimisation28 using static trial data for model calibration, to determine muscle and joint forces for each sampled frame. The objective function applied was:
where Fi is the force output of the ith muscle element, \(F_{{i}_{\hbox{max}}}\) defines the ith muscle element’s force at maximum isometric contraction and n is the total number of muscle elements − 163.26\(F_{{i}_{\hbox{max}}}\) was calculated for each element from physiological cross-sectional area, which was determined using subject-specific measurements and anatomical values obtained from the same matched-subject MRI described above.
To formulate the targets for training, the resulting scalar values of each timestep were combined into individual vectors, for each trial, of dimensions [t, 1]. Again, many such vectors from different trials were concatenated to form a single long target vector corresponding to the input data matrix.
For EMG signal prediction, the raw signal was full-wave rectified, smoothed and resampled to match the length of the input tensor. Root mean square smoothing6 was performed using a window size of 50 ms. All values were normalised to the observed within-trial maximum, producing activation trajectories that varied between zero and one for each muscle in every trial.
Training/Validation/Test Split
The data were randomly separated into three groups according to an 80/10/10 relative split for the purposes of training, validation and final evaluation. Two types of final testing were performed. First, data obtained from individual subjects were strictly isolated within one group only to allow extrapolation of test evaluation metrics to new subjects (‘subject-naive’ setting). This was achieved by randomisation at the subject level. In the second, ‘subject-exposed’ setting, for every trial in the test set, there was at least one other trial from the same subject included in the training set. This was achieved by active redistribution of trial data following randomisation, where required, from the test set to the training set. The loss function for network training was defined as the root mean square error between predicted and actual target values:
where \(y_{i}\) represents the target value of the ith training example, generated by Freebody inverse dynamics for a given trial at time ti, \(x_{i}\) is the corresponding input tensor at ti and \(\phi\) represents the transform function applied by the neural network. Training was performed using the backpropagation algorithm32 and network weights optimised using Adam.17 Final training time was roughly one hour (NVIDIA Titan Xp GPU).
Neural Network Architecture
The validation data were used to optimise network architecture and hyperparameters for the prediction task. Simple feedforward, convolutional and recurrent architectures were tested, and it was established that validation accuracy was greatest using a convolutional neural network with fully connected output layers so hyperparameter searches were henceforth focussed here. In order to facilitate 2-dimensional convolution, the input at each timestep was reshaped to dimensions (x, 3), that is, each row of the input matrix corresponded to the 3-dimensional position vector of a single marker, or the 3-dimensional vectors of the ground reaction force or centre of pressure. Batch normalisation12 was found to improve accuracy and speed of convergence, and was used in all tested networks, applied as per convention to all layers bar the output. All networks utilised ReLU activations in their hidden layers, with a single linear output neuron. Manual hyperparameter optimisation was performed incrementally to establish reasonable bounds for subsequent search. Random sweeps within these bounds were then performed to find optimum architecture and hyperparameters. A single network architecture was developed and optimised for all predictions, and validation of the final network was performed in two ways.
Validation 1: Comparison with Inverse Dynamics and Static Optimisation/EMG Data
Final evaluation was performed for the prediction of major muscle group force magnitudes, EMG activations and the medial knee joint reaction force. Particular emphasis was given to the latter because of the high level of validation accuracy that has been achieved by musculoskeletal models for its prediction, and because of its established clinical relevance as a marker for the risks of onset and progression of OA of the knee.25,35 Quantitative evaluation of accuracy was performed by comparison of the neural network’s predictions with those obtained when the same input data were analysed by inverse dynamic modelling, or with the true EMG signal.
Validation 2: Grand Challenge Data
A neural network was trained to predict medial knee contact force in each of the grand challenge competition years. Input data were restricted to include only the three-dimensional ground reaction force vector; this and instrumented prosthesis data were resampled in order to equate lengths. Network architecture was identical to that previously used for the prediction of model force outputs, but potent regularisation techniques were necessary given the relative paucity of data available – these included L2 weight decay and early stopping. A separate model was trained for each year of the competition, by training using the data from all of the other years, a subset of which was withheld for the purposes of validation. Final evaluation was performed on the two trials that made up the evaluation trials in the real Grand Challenge of that particular year. The remainder of that year’s data was discarded from both training and test sets in order to match the data available to Grand challenge competitors as closely as possible, for the purposes of fair comparison. Rotation of the test dataset (leave-one-out cross-validation) yielded accuracy metrics for each of the six competition years.
Real time Implementation
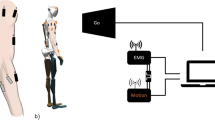
For real time streaming of kinematic and kinetic data direct to the gait lab console, the Vicon SDK plugin38 was used and adapted to incorporate the trained neural network. Input kinematic and kinetic data were processed in real time and forward-propagated through the network, returning values for internal force magnitudes. A custom plotting interface was developed in C++ for real time visualisation.
Network Interrogation
A neural network was trained to predict the medial knee joint reaction force taking 3-dimensional intersegmental angle data and ground reaction force as input. Following completion of network training, examples were individually forward-propagated through the network and at each time step the partial derivatives of the output with respect to input elements were computed. The gradients thus obtained per trial were resampled from trial length to length 100 to allow for cross comparison and averaging across trials. Average values were multiplied by the standard deviation across trials of each feature at each time step in order to obtain a measure of the relative contribution of each feature to the variance of the medial knee JRF across stance phase.
Results
555 trials were successfully processed using Freebody inverse dynamics. A small number of trials were discarded because they failed to yield feasible solutions to optimisation. The data were used to train a number of different neural networks implemented in Python using the Tensorflow library for numerical computation.1
Network Architecture
Optimum architecture and parameters are illustrated in Figure 1 for a network taking the full extent of kinematic and kinetic data as input. All convolutional layers were 2-dimensional, applying a kernel size of [1× 3] with stride size 1. To ensure equivalence of the first two data dimensions, the input was padded with zeros prior to application of the convolutional operator, as per convention.
Validation 1: Accuracy vs. Inverse Dynamics/Static Optimisation and EMG
The neural network outputs were compared with those of Freebody and EMG sensor data as appropriate. Neural networks that had been exposed to other data of the test subject during training (‘subject-exposed’, below) in the majority of cases outperformed those that had not.
Medial knee JRF Prediction
See Table 2 and Figs. 2a, 2b, 3, and 4
Muscle Force Prediction
See Table 3 and Figs. 5a and 5b
EMG prediction
See Table 4.
Computation Time (Average Per Trial)
Average computational time required for inference was 71 milliseconds per trial, compared with 16 minutes for Freebody inverse dynamics and static optimisation (Intel Core i7 6700).
Validation 2: Grand Challenge Validation
Neural network outputs were compared with ground truth tibiofemoral force data obtained from instrumented prostheses, as in the real Grand challenges. Evaluation metrics bettered those of the winning submissions in 4 of the 6 competitions (Table 5).
Real Time Implementation
Real time deployment to the gait lab was successful, with operation speeds in excess of 100Hz for the prediction of internal forces from force plate data.
Network Interrogation
Feature importance maps derived by interrogation of a neural network trained on the entirety of the data showed smooth variation over stance phase (Fig. 6).
Discussion
Supervised learning was proposed many decades ago as a suitable framework for the posing of the motor control problem. Since then, deep neural networks have enjoyed a resurgence of popularity, driven by a number of impressive performances in difficult problems including computer vision and speech recognition. More recently there have been attempts to fuse the disparate areas of deep learning and neuroscience.22
Here, a novel integration of deep learning with musculoskeletal modelling was used to demonstrate advantages from the use of supervised learning techniques in approximating the mapping from kinematic space to muscle space. Good accuracy in force prediction was achieved across a diverse test cohort, with error metrics falling within the bounds of variability resulting from the application of different anatomical datasets, for example.9 Performance in subjects with pathology was, as expected, slightly reduced in most cases relative to the entire test population, likely reflecting a greater variability in kinematic and kinetic features, and EMG signals. Some disparity in accuracy was generally observed between subject-exposed and subject-naïve models, for prediction of both inverse dynamic outputs and the EMG signal. This likely reflects the relatively small size of the training dataset, so that similarity of the train and test distributions was not guaranteed in the subject-naïve case. Until sufficient data are available from a broad range of subjects, the use of subject-specific models will be necessary if the greatest levels of accuracy are required. A major advantage of supervised learning over physics-based modelling is speed; this enabled the development of a real time lab-based system for feedback of internal forces.
Comparison with Existing Real Time Methods
Real time feedback is not new27,37 but until now true real time rates have often required a number of compromises in terms of the complexity of the employed modelling. In contrast, the approach used here, by bypassing the computationally demanding explicit execution of inverse kinematics, inverse dynamics and static optimisation, promises the ability to build networks based on the results of models of increasing complexity, including those that so far have failed to achieve widespread use in large part because of their intractability to computation, such as those based on dynamic optimisation.3 Though obtaining data using such complex models would be computationally expensive, the costs of network training and inference are not expected to differ significantly from those reported here. In fact, the computational costs of inference are low enough to make the deployment of trained networks to cheap portable hardware, including mobile phones, feasible, raising the prospect of force estimation away from the confines of the gait laboratory.
A further contribution of the work is in the development of means for musculoskeletal force prediction in the absence of the full ensemble of input data required for conventional modelling. Though accuracy was greatest when the neural network was exposed to all of the input data, reasonable performance was attained with only part of it, suggesting a degree of redundancy of information in the inputs. Machine learning techniques should provide a natural means for the exploitation of such redundancy. To test this hypothesis, validation on Grand Challenge data was performed using the three-dimensional ground reaction force only as input, and the resulting models were competitive with and, in most cases, bettered the winning submissions produced using detailed, handcrafted musculoskeletal models that utilised all of the available input data. This lessening of the input data requirement is particularly important in the clinical domain, where time and/ or resource constraints may be limiting. In the same vein, and in stark contrast to physics-based modelling, the network-driven approach requires no input of body segment parameters and no calibration trials, further increasing its attractiveness.
The applications of the technique are broad. True real time operation enables the use of model outputs as biofeedback for the training of movement parameters. For subjects with OA, an important application is gait retraining with the purpose of reducing the medial knee joint reaction force. It has been shown that real time biofeedback can be used to modify important kinetic parameters in healthy subjects,40 but as yet the difficulty of true force prediction has meant that this has not been accomplished for the medial knee JRF in patients.
Reasonable accuracy was obtained for prediction of the EMG signal from some muscles of the lower limb, but for others significant variability in the signals recorded from different trials of different subjects meant that error metrics were relatively high. A lack of repeatability is often a problem with the standard use of real EMG signals,30 as inter-trial differences, for example in electrode placement, may have major effects on the results obtained. There are other issues with the standard use of EMG, including a requirement for expertise both in the acquisition and the interpretation of sensor data, and, of course, the requirement for the sensors themselves, which may be expensive. For subjects who are sufficiently similar to those upon whose data supervised models have been built, or better yet where models have been exposed to previous trials of a given subject, such models can provide predictions of the EMG signal quickly and without extensive pre-processing of input data.
Calculation of partial derivatives for the joint reaction force with respect to input joint angles allowed for the generation of feature importance maps illustrating a measure of the importance of features in the input for the prediction of the trained objective. To our knowledge, this represents the first application of the technique to motion data. These maps showed smooth variation in importance across stance phase, with several features inverting the sign of their contribution to the medial knee JRF. The contributions of two components, the knee abduction angle and the vertical component of the ground reaction force, fit with the well-established correlation between the external knee adduction moment, with which these two factors are themselves strongly correlated, and the medial knee joint reaction force. Incidentally, recent work supports the reliability of cohort-level modelling over that performed on the basis of measurements from individual subjects.29
It has been demonstrated that neural networks are able to generalise well to subjects of varying body morphology, age and pathology, often with very different gait patterns. However, this was shown only for a single activity; confidence in the model’s ability to provide accurate predictions for other types of movement remains unwarranted. Moreover, in the clinical domain, it is likely that rather than a capacity for accuracy in prediction of absolute force values, the ability to reveal trends in musculoskeletal forces with, for example, gait modification, will be of greater utility. Again, as yet, this ability remains unproven. Greatest accuracy will be achieved with the training of subject specific models, with the inclusion of a wide range of activities in the training set.
The requirement for the use of conventional musculoskeletal models to generate training data stems from a lack of sufficient available data on internal forces from other sources, a consequence of the difficulty of obtaining measures by other means, such as instrumented prostheses. However, the use of these models is predicated on the assumptions that underlie the solution of the muscle redundancy problem, and modelling is also subject to errors arising from experimental techniques such as motion capture. One of the perceived benefits of supervised learning is that it affords the ability to depart from the conventional framework, and, in doing so, leave behind some of these disadvantages. Training using data available from the Grand Challenges showed that, contrary to popular conception, neural networks can compete with and often, in fact, outperform meticulously handcrafted solutions where only limited amounts of training data are available. In future, as more sensor data become available, the potential to use supervised learning to obtain accurate predictions, and the conditions under which it is possible to do so, are expected to grow.
The use of neural networks to compute the mapping from the kinematic space to force space brings several advantages to musculoskeletal force prediction. Ultimately, it is hoped that this new technique will be useful in the clinical domain. A promising application is that of real time gait analysis and feedback, where the benefit of speed is of particular utility.
References
Abadi, M., et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:160304467, 2016.
Albus, J. S. A theory of cerebellar function. Math. Biosci. 10(1–2):25–61, 1971.
Anderson, F. C., and M. G. Pandy. Dynamic optimization of human walking. J. Biomech. Eng. 123:381–390, 2001.
Ardestani, M. M., M. Moazen, Z. Chen, J. Zhang, and Z. Jin. A real-time topography of maximum contact pressure distribution at medial tibiofemoral knee implant during gait: application to knee rehabilitation. Neurocomputing 154:174–188, 2015.
Cleather, D. J., and A. M. J. Bull. The development of a segment-based musculoskeletal model of the lower limb: introducing Freebody. R. Soc. Open Sci. 2(6):140449, 2015.
De Luca, C. J. The use of surface electromyography in biomechanics. J. Appl. Biomech. 13(2):135–163, 1997.
Ding, Z., D. Nolte, C. K. Tsang, D. J. Cleather, A. E. Kedgley, and A. M. J. Bull. In vivo knee contact force prediction using patient-specific musculoskeletal geometry in a segment-based computational model. J. Biomech. Eng. 138(2):021018, 2016.
Eskinazi, I., and B. J. Fregly. Surrogate modeling of deformable joint contact using artificial neural networks. Med. Eng. Phys. 37(9):885–891, 2015.
Gerus, P., et al. Subject-specific knee joint geometry improves predictions of medial tibiofemoral contact forces. J. Biomech. 46(16):2778–2786, 2013.
Heller, B. W., P. H. Veltink, N. J. Rijkhoff, W. L. Rutten, and B. J. Andrews. Reconstructing muscle activation during normal walking: a comparison of symbolic and connectionist machine learning techniques. Biol. Cybern. 69(4):327–335, 1993.
Hinton, G., et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process. Mag. 29(6):82–97, 2012.
Ioffe S., and C. Szegedy. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167, 2015.
Kallweit, S., T. Müller, B. McWilliams, M. Gross, and J. Novák. Deep Scattering: Rendering Atmospheric Clouds with Radiance-Predicting Neural Networks. arXiv:170905418, 2017.
Kannape, O. A., and O. Blanke. Self in motion: sensorimotor and cognitive mechanisms in gait agency. J. Neurophysiol. 110(8):1837–1847, 2013.
Kawato, M., K. Furukawa, and R. Suzuki. A hierarchical neural-network model for control and learning of voluntary movement. Biol. Cybern. 57(3):169–185, 1987.
Kim, H. J., J. W. Fernandez, M. Akbarshahi, J. P. Walter, B. J. Fregly, and M. G. Pandy. Evaluation of predicted knee-joint muscle forces during gait using an instrumented knee implant. J. Orthop. Res. 27(10):1326–1331, 2009.
Kingma, D., and J. Ba. Adam: A method for stochastic optimization. arXiv:14126980, 2014.
Kinney, A. L., T. F. Besier, D. D. D’Lima, and B. J. Fregly. Update on grand challenge competition to predict in vivo knee loads. J. Biomech. Eng. 135(2):021012, 2013.
Krizhevsky, A., I. Sutskever, and G. E. Hinton. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 1097–1105, 2012.
Liu, M. M., W. Herzog, and H. H. Savelberg. Dynamic muscle force predictions from EMG: an artificial neural network approach. J. Electromyogr. Kinesiol. 9(6):391–400, 1999.
Long, M. J., E. Papi, L. D. Duffell, and A. H. McGregor. Predicting knee osteoarthritis risk in injured populations. Clin. Biomech. 47:87–95, 2017.
Marblestone, A. H., G. Wayne, and K. P. Kording. Toward an integration of deep learning and neuroscience. Front. Comput. Neurosci. 10:94, 2016.
Marr, D. A theory of cerebellar cortex. J. Physiol. 202(2):437–470, 1969.
Mehrizi, R., X. Peng, Z. Tang, X. Xu, D. Metaxas, and K. Li. Toward Marker-free 3D pose estimation in lifting: a deep multi-view solution. 13th IEEE International Conference on Automatic Face & Gesture Recognition, pp. 485–491, 2018.
Miyazaki, T., M. Wada, H. Kawahara, M. Sato, H. Baba, and S. Shimada. Dynamic load at baseline can predict radiographic disease progression in medial compartment knee osteoarthritis. Ann. Rheum. Dis. 61:617–622, 2002.
Modenese, L., A. T. M. Phillips, and A. M. J. Bull. An open source lower limb model: hip joint validation. J. Biomech. 44:2185–2193, 2011.
Pizzolato, C., M. Reggiani, D. J. Saxby, E. Ceseracciu, L. Modenese, and D. G. Lloyd. Biofeedback for gait retraining based on real-time estimation of tibiofemoral joint contact forces. IEEE Trans. Neural Syst. Rehabil. Eng. 25(9):1612–1621, 2017.
Potra, F. A., and S. J. Wright. Interior-point methods. J. Comput. Appl. Math. 124(1):281–302, 2000.
Price, P. D. B., C. Gissane, and D. J. Cleather. Reliability and minimal detectable change values for predictions of knee forces during gait and stair ascent derived from the freebody musculoskeletal model of the lower limb. Front. Bioeng. Biotechnol. 5:74, 2017.
Reaz, M. B., M. S. Hussain, and F. Mohd-Yasin. Techniques of EMG signal analysis: detection, processing, classification and applications. Biol. Proced. Online 8(1):11, 2006.
Rodacki, A. L. F., N. E. Fowler, and S. J. Bennett. Vertical jump coordination: fatigue effects. Med. Sci. Sports Exerc. 34:105–116, 2002.
Rumelhart, D. E., G. E. Hinton, and R. J. Williams. Learning representations by back-propagating errors. Nature 323(6088):533–536, 1986.
Sepulveda, F., D. M. Wells, and C. L. Vaughan. A neural network representation of electromyography and joint dynamics in human gait. J. Biomech. 26(2):101–109, 1993.
Shadmehr, R., and F. A. Mussa-Ivaldi. Adaptive representation of dynamics during learning of a motor task. J. Neurosci. 14(5 Pt 2):3208–3224, 1994.
Sharma, L., et al. Knee adduction moment, serum hyaluronan level, and disease severity in medial tibiofemoral osteoarthritis. Arthritis Rheum. 41:1233–1240, 1998.
Trepczynski, A., I. Kutzner, G. Bergmann, W. R. Taylor, and M. O. Heller. Modulation of the relationship between external knee adduction moments and medial joint contact forces across subjects and activities. Arthritis Rheum. 66:1218–1227, 2014.
van den Bogert, A. J., T. Geijtenbeek, O. Even-Zohar, F. Steenbrink, and E. C. Hardin. A real-time system for biomechanical analysis of human movement and muscle function. Med. Biol. Eng. Comput. 51(10):1069–1077, 2013.
Vicon. Datastream SDK. 1.7.0 ed. https://www.vicon.com/products/software/datastream-sdk2013.
Wallace, I. J., S. Worthington, D. T. Felson, R. D. Jurmain, K. T. Wren, H. Maijanen, R. J. Woods, and D. E. Lieberman. Knee osteoarthritis has doubled in prevalence since the mid-20th century. Proc. Natl. Acad. Sci. USA 114(35):9332–9336, 2017.
Wheeler, J. W., P. B. Shull, and T. F. Besier. Real-time knee adduction moment feedback for gait retraining through visual and tactile displays. J. Biomech. Eng. 133:041007, 2011.
Wolpert, D. M., R. C. Miall, and M. Kawato. Internal models in the cerebellum. Trends Cogn. Sci. 2(9):338–347, 1998.
Xia, P., J. Hu, and Y. Peng. EMG-based estimation of limb movement using deep learning with recurrent convolutional neural networks. Artif. Organs 42(5):E67–E77, 2018.
Yamashita, Y., and J. Tani. Emergence of functional hierarchy in a multiple timescale neural network model: a humanoid robot experiment. PLoS Comput. Biol. 4(11):e1000220, 2008.
Acknowledgments
The dataset analysed during the current study was acquired through funding from the Engineering and Physical Sciences Research Council and the Wellcome Trust as part of the Medical Engineering Solutions in Osteoarthritis Centre of Excellence at Imperial College London. We gratefully acknowledge the support of NVIDIA Corporation for the donation of the Titan Xp GPU used for this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Estefanía Peña oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rane, L., Ding, Z., McGregor, A.H. et al. Deep Learning for Musculoskeletal Force Prediction. Ann Biomed Eng 47, 778–789 (2019). https://doi.org/10.1007/s10439-018-02190-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-018-02190-0