Abstract
The Greenlandic and west-central Siberian breeding populations of Sanderlings Calidris alba are separated by ca. 2000 km during the breeding season, but mix in Europe to some extent during migration. However, the number of Siberian Sanderlings that spend the non-breeding season along the East Atlantic Flyway (extending from western Europe to South Africa), if any, is unknown. Although both populations are considered part of the nominate subspecies C. a. alba based on morphology, population structure in Sanderlings has yet to be described with molecular methods. We examined genetic differentiation at the mtDNA control region (CR) and seven microsatellite loci between Greenland- and Siberia-breeding Sanderlings in order to: (1) develop a diagnostic tool for assessing the breeding origin of Sanderlings along the East Atlantic Flyway, and (2) provide a comparison with the co-distributed and ecologically similar Red Knot, in which CR differentiation of geographically analogous populations (C. canutus islandica and C. c. canutus) has indicated isolation of lineages near the time of the last glacial maximum. By contrast, we found only weak differentiation between the Sanderling breeding populations at the CR, and no differentiation at microsatellite loci. These results suggest that the assignment of breeding origin of Sanderlings on Afro-European flyways will not be possible with simple and inexpensive genetic methods, and imply that Sanderlings and Red Knots have very different post-glacial phylogeographic histories.
Zusammenfassung
Geringe genetische Unterschiede zwischen grönländischen und sibirischen Sanderling-Populationen ( Calidris alba ) weisen auf eine andere phylogeographischen Herkunft als die des Knutt ( Calidris canutus ) hin
Die grönländischen und die west-zentral-sibirischen Brutpopulationen des Sanderlings (Calidris alba) sind während der Brutzeit ca. 2.000 km voneinander entfernt, mischen sich aber zu einem guten Teil während der Zugzeit. Allerdings ist die Anzahl derjenigen sibirischen Sanderlinge, die sich außerhalb der Brutzeiten entlang der ostatlantischen Zugroute (reicht von Westeuropa bis nach Südafrika) aufhalten, nicht bekannt. Obwohl beide Populationen aufgrund ihrer Morphologie als Teile der Nominalform Calidris alba alba angesehen werden, steht für den Sanderling eine Beschreibung der Populations-Struktur mit molekularbiologischen Methoden noch aus. Wir bestimmten für die mtDNA Kontrollregion (CR) und für sieben Mikrosatellitenloci die genetischen Unterschiede zwischen in Grönland und in Sibirien brütenden Sanderlingen, (1) um ein Diagnose-Werkzeug zu entwickeln, mit dem der Ursprung von Sanderlingen entlang der ostatlantischen Zugroute bestimmt werden kann, und (2) um einen Vergleich mit dem verbreitungsmäßig und ökologisch ähnlichen Knutt zu ermöglichen, für die eine CR-Differenzierung der geographisch analogen Populationen (C. canutus islandica and C. c. canutus) die Aufspaltung der Abstammungslinien etwa zur Zeit des letzteiszeitlichen Maximums andeutet. Stattdessen fanden wir auf der CR nur schwache Unterschiede zwischen den Brutpopulationen des Sanderlings und gar keine Unterschiede bei den Mikrosatellitenloci. Diese Ergebnisse legen nahe, dass eine Zuordnung der Herkunftsgegend von Sanderlingen auf der afrikanisch-europäischen Zugroute nicht mit einfachen und preiswerten genetischen Methoden möglich sein wird und dass die phylogeographische Entwicklung von Sanderling und Knutt nach der Eiszeit sehr unterschiedlich verlaufen ist.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the East Atlantic Flyway, the non-breeding range of Sanderlings Calidris alba includes the British Isles and extends nearly the entire Atlantic coast from the North Sea to the southernmost tip of Africa (Scott 2009). However, the breeding origin of much of this non-breeding population, estimated to be 123,000 individuals (Stroud et al. 2004), remains in question (Reneerkens et al. 2009). It was formerly believed that Sanderlings wintering in western Europe were of Siberian breeding origin, whereas those breeding in northeast Greenland appeared in western Europe only during migration to and from non-breeding areas in western Africa (Green and Greenwood 1978; Langston 2002; Meltofte et al. 1994). This view largely originated from the observation of two distinct cohorts of Sanderlings in the United Kingdom: one that performed pre-basic moult and remained there for the winter, and another that appeared only during autumn and spring migrations and did not moult locally (Green and Greenwood 1978).
More recently, an increasing number of individual ring-recoveries have partly elucidated migratory patterns of Sanderlings on the East Atlantic and West Asian-East African Flyways. Based on birds ringed during breeding in northeast Greenland or on passage in Iceland, Greenland-breeding Sanderlings use non-breeding areas along the North Sea and Atlantic coasts from the British Isles to South Africa (Gudmundsson and Lindström 1992; Reneerkens et al. 2009), and include both European cohorts (i.e., moulting residents and non-moulting passage birds; Reneerkens et al. unpubl. data). In addition, at least some birds breeding on Ellesmere Island, Canada migrate to western Europe (Reneerkens et al. 2008). Meanwhile, part of the central Siberian-breeding population appears to follow a loop-migration route southward through the Caspian and Black Sea regions to eastern and southern Africa, and then northward via western Africa, the Mediterranean, and western Europe; this has been largely derived from birds ringed either in South Africa or during northward migration in Europe (Underhill et al. 1999). To date, there has been no unequivocal evidence of Siberian Sanderlings spending the non-breeding season in western Africa or Europe (Reneerkens et al. 2009); however, due to the low numbers of individuals ringed or recovered in Siberia, this possibility cannot currently be ruled out. Therefore, although the non-breeding ranges of the Siberian and Greenlandic breeding populations clearly overlap in South Africa (Underhill et al. 1999), the extent of mixing in Europe and western Africa outside the migratory periods is unknown. This uncertainty limits our ability to identify population trends on either flyway or to link breeding conditions or performance with patterns in the non-breeding season.
Globally, two subspecies of Sanderling have been recognized based on small differences in morphometrics and breeding plumage (Engelmoer and Roselaar 1998): C. a. alba breeds in northeastern Greenland, Ellesmere Island, Svalbard, and central Siberia, whereas C. a. rubidus breeds in northern Alaska and the central Canadian Arctic (Engelmoer and Roselaar 1998; but see Tomkovich and Serra 1999). Therefore, all Sanderlings on the East Atlantic and West Asian-East African Flyways are currently considered part of the nominate subspecies C. a. alba, despite their use of multiple geographically isolated breeding areas and distinct migration routes. However, genetic population structure across flyways has yet to be investigated in Sanderlings.
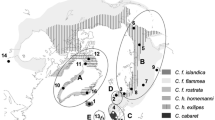
Sanderlings and Red Knots Calidris canutus are ecologically similar congeners that use predominantly high-Arctic tundra breeding habitats (latitudes 63°–80°), and their largely overlapping global breeding distributions (Fig. 1) may have developed contemporaneously through similar population responses to post-glacial changes in habitat and ecology (e.g., Gilg and Yoccoz 2010). Based on population structure detected in mitochondrial DNA (mtDNA) in Red Knots, Buehler et al. (2006) hypothesized a global expansion from two Palearctic refugia present at the Last Glacial Maximum (LGM). An unexpected finding was that C. c. canutus (breeding in central Siberia) and C. c. islandica (breeding in Greenland and northeastern Canada), previously considered sister taxa based on morphological similarities, were actually one of the most distantly related pairs (F ST = 0.19) among six subspecies (pairwise F ST range = 0.002–0.27) and the most differentiated among adjacent flyway pairs (F ST range = 0.002–0.19; Buehler and Baker 2005). The authors proposed that, after diverging approximately 23,000 years ago, Nearctic and Palearctic lineages came into secondary contact when Red Knots established a new migratory route from the Canadian Arctic to Europe, perhaps as recently as 1000 years ago (Buehler et al. 2006).
a Global breeding distribution of Sanderling (in dark gray). Heavy dotted lines indicate presumed boundaries between subspecies C. a. alba and C. a. rubidus based on morphology (Engelmoer and Roselaar 1998). Circles indicate origins of samples in this study. b Global breeding distribution of Red Knot (in dark gray). Heavy dotted lines indicate presumed boundaries between Nearctic and Palearctic clades based on population structure in mtDNA (Buehler and Baker 2005). Range information adapted from BirdLife International (2013) and Lappo et al. (2012)
The central Siberian and Greenlandic populations of Sanderling are, respectively, close analogues of C. c. canutus and C. c. islandica in terms of breeding distribution and migration routes; both population pairs are separated by approximately 2000 km in the breeding season but meet in Europe during migration and show some extent of overlap in the non-breeding season. If the Sanderling populations are genetically differentiated to a similar degree as their Red Knot counterparts, this may: (1) provide a diagnostic tool for assessing the breeding origin of Sanderlings along the East Atlantic Flyway, and (2) support a similar recent evolutionary history and global expansion in the two species. Therefore, we examined genetic differentiation at the mtDNA control region and seven microsatellite loci between Sanderlings breeding in northeast Greenland and the Taimyr Peninsula of central Siberia.
Methods
Sample origins
All samples were collected at known breeding areas in northeast Greenland or the Taimyr Peninsula of central Siberia. In June–August 1990/1992, tissue samples of eight adults (five males, three females) and one juvenile female were collected at Knipovich Bay, Russia (north-central Taimyr Peninsula; 76.1°N, 98.5°E; see Soloviev and Tomkovich 1995). In July 1994, blood samples of three adults (one male, two females) were collected at Cape Sterlegova, Russia (northwest Taimyr Peninsula; 75.4°N, 89.1°E; see Tulp et al. 1998). In July–July 2003/2007, blood samples of 12 adults (6 males, 6 females) were collected at Zackenberg Research Station, Greenland (74.5°N, 21.0°W; see Meltofte and Rasch 2008).
Sequencing and genotyping
We extracted total cellular DNA with the Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s instructions. Extracted DNA was stored in TE buffer (10 mM Tris–HCl [pH 8.0], 1 mM EDTA) at −20 °C. We determined the sex of individuals following Fridolfsson and Ellegren (1999) and using calidrid-specific primers 2602F and 2669R (Reneerkens et al. 2014).
We amplified the 5′ end of the mtDNA control region (CR) using primers L98F and H772R (Wenink et al. 1993). The PCR profile consisted of an initial denaturation of 2 min at 94 °C, followed by 25 cycles of 30 s at 94 °C, 30 s annealing at 54 or 58 °C, and an extension of 2 min at 72 °C. The final concentrations in the 10 μL PCR (including 1 μL DNA template) were 1 μM of each primer, 1× Taq DNA polymerase buffer, 3.2 mM dNTPs, and 0.03 U/μL Taq DNA polymerase (Invitrogen, Inc.). PCR products and negative controls were checked by electrophoresis through a 2 % agarose gel. PCR products were enzymatically cleaned (following Werle et al. 1994), sequenced in both directions using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc.) according to manufacturer's instructions, and analyzed on an ABI 3730 DNA Analyzer (Applied Biosystems, Inc.). Sequences were aligned and edited in Geneious Pro ver. 5.5.5 (Biomatters Ltd.).
We genotyped individuals at seven microsatellite loci, including five developed specifically for Sanderlings (an3, gt24, m11, m14, m18; Luttikhuizen et al. 2011) and two originally identified in Pectoral Sandpiper Calidris melanotus and also polymorphic in Sanderlings (cme1, cme6; Carter and Kempenaers 2007; Luttikhuizen et al. 2011). PCR profiles varied by locus; for gt24 and m11: denaturation of 4 min at 94 °C; 32 cycles of 30 s at 94 °C, 30 s annealing at 54 °C, and extension of 15 s at 72 °C; plus a final extension of 10 min at 72 °C. For the remaining loci: denaturation of 2 min at 94 °C; 15 cycles of 30 s at 94 °C, 90 s annealing at 56 °C, and extension of 60 s at 72 °C; 15 (cme1, cme6, m14) or 20 (an3, m18) cycles of 30 s at 94 °C, 90 s annealing at 60 °C, and extension of 60 s at 72 °C; plus a final extension of 30 min at 60 °C. The total reaction volumes were 10 μL, containing 1.0 μL DNA, 1× Taq buffer, 3.2 mM dNTPs, 0.04 U/μL Taq DNA polymerase (Invitrogen, Inc.), and 1 μM of each primer, fluorescently labeled to enable pooling of loci. Fragment analyses were performed on an ABI 3730 DNA Analyzer with GeneScan 500 ROX Size Standard (Applied Biosystems, Inc.). Allele sizes were assigned using Genemapper ver. 4.0 (Applied Biosystems, Inc.) and then converted to the number of repeats (using repeat motifs determined by Carter and Kempenaers 2007; Luttikhuizen et al. 2011).
Data analysis
For mtDNA, we used DNAsp ver. 5.10 (Librado and Rozas 2009) to estimate haplotype and nucleotide diversity, and Arlequin ver. 3.5.1 (Excoffier et al. 2005) to assess deviations from mutation-drift equilibrium indicating changes in population size (Tajima’s D) and to estimate degree of population differentiation, according to both haplotype frequencies (F ST) and genetic distance (φ ST). The P values were estimated from 10,000 permutations. We estimated the haplotype network in Network ver. 4.6.1 (Fluxus Technology Ltd.). For microsatellite loci, we used Arlequin ver. 3.5.1 to estimate population differentiation (distance method; φ ST), and GenePop ver. 4.2 (Raymond and Rousset 1995) to assess heterozygosity, deviations from expected Hardy–Weinberg genotype frequencies, and linkage disequilibrium. To minimize Type I errors in multiple comparisons, we controlled the false discovery rate using the step-up method of Benjamini and Hochberg (1995).
To investigate whether low-level population differentiation was associated with historical isolation followed by homogenizing gene flow or recent isolation, we employed coalescent analysis in IMa2 (Hey and Nielsen 2004) to derive effective population size (N e, from estimates of θ), migration rates (N m , from m), and divergence time (T div, from t). For both mtDNA and microsatellite loci, we compared two models: (1) asymmetrical migration, and (2) no migration. In a series of trial runs, we adjusted prior ranges to best achieve unimodal posterior distributions; we assessed convergence by inspection of the parameter trend plots. We ensured independence of runs by adjusting heating terms to achieve swapping rates consistently greater than 0.60. After a burn-in of 10,000 steps, the parameter space was sampled by 200 adaptively heated chains, sampling 40,000 trees, and 100 independent replicate runs with different random seeds, generating a total of four million genealogies in IMa2 M-mode. Subsequently, parameters estimates were generated by randomly sampling 300,000 genealogies across the 100 runs (IMa2 L-mode). For calculations of T div and N e for mtDNA, we adopted the CR mutation rate of 6.96 × 10−8 substitutions/site/year used for Red Knots by Buehler and Baker (2005). For microsatellites, we used a general vertebrate mutation rate of 1 × 10−4 substitutions/locus/generation (Whittaker et al. 2003); note that microsatellite characteristics related to mutation rate such as length, allele dispersion, and allele size range do not differ between birds and mammals (Neff and Gross 2001). We assumed a generation time (age at first reproduction) of two years (Macwhirter et al. 2002).
Results
Population differentiation
We obtained a 508-bp segment of mtDNA CR domains I and II for 12 individuals from Greenland and 11 from Siberia; sequencing failed for one Siberian sample. Among the 23 mtDNA sequences, we detected nine segregating sites, resulting in a total of 12 unique haplotypes (Fig. 2; GenBank accession numbers KT594771–KT594782). However, no segregating sites were diagnostic for population. Three haplotypes appeared in both populations, and the nine remaining haplotypes were identified only in single individuals. Haplotype diversity was 0.872 for Siberia, 0.848 for Greenland, and 0.870 overall. Nucleotide diversity was low: 0.0033 ± 0.0007 for Siberia, 0.0031 ± 0.0005 for Greenland, and 0.0032 ± 0.0007 overall. In mtDNA, we found a moderate but statistically non-significant degree of population differentiation according to haplotype frequencies (F ST = 0.061, P = 0.089) but no differentiation by the distance method (φ ST = –0.0017, P = 0.39). We found some indication of population expansion, albeit statistically non-significant (D = −0.73, P = 0.25).
We successfully genotyped all 24 individuals at five microsatellite loci (an3, cme1, cme6, gt24, and m18); for two additional loci, we removed some individuals due to ambiguous genotypes (one Siberian and three Greenlandic individuals for locus m11, and one Greenlandic individual for m14). We detected no significant departure from linkage equilibrium (Fisher’s exact test; all locus pairs: P = 0.14–1.00). After correcting for multiple comparisons, we detected no significant locus-specific deviations from Hardy–Weinberg proportions, with populations considered either singly or pooled (Table 1); therefore, there was no evidence of excess homozygosity related to null alleles or homoplasy. We detected no population differentiation at microsatellite loci (φ ST = −0.0050, P = 0.69).
Divergence scenarios
Given the geographic isolation of the two breeding ranges, the weak differentiation we observed could result from either historical isolation followed by significant gene flow, or recent isolation with little or no subsequent gene flow. We could only partly resolve this question with coalescent analysis in IMa2, because for both mtDNA and microsatellites, neither model 1 (allowing asymmetrical migration between Siberia and Greenland) nor model 2 (no migration) fully converged for all parameter estimates. Specifically, we could not estimate migration rates or population-specific N e because we failed to achieve complete unimodal distributions for θ 1, θ 2, m 1, or m 2. Therefore, we only calculated estimates for T div and ancestral N e from each model; within each model, these parameter estimates were robust in trial runs to a wide variety of prior ranges in all parameters (not shown). All models indicated a recent divergence: estimates of T div were all <10,000 years, with 95 % CI boundaries including 0–21,000 years (Table 2).
Discussion
We found no clear genetic differentiation between two Sanderling breeding populations separated by approximately 2000 km. Although we detected a non-trivial, but non-significant, degree of differentiation in mtDNA haplotype frequencies, the complete lack of differentiation shown by genetic distance in mtDNA (reflected in Fig. 2) and microsatellites implies either significant gene flow between Greenlandic and Siberian populations or recent isolation that is yet to resolve into distinct lineages. The absence of strong deviations from Hardy–Weinberg proportions (which should arise with non-random mating) when microsatellite loci were pooled across populations (Table 1), further attests to a lack of structure. Our results demonstrate that simple and inexpensive genetic methods will not be useful for determining the breeding origin of Sanderlings on the East Atlantic and West Asian-East African Flyways, whether by directly assigning individuals to breeding region (e.g., Lopes et al. 2013), or by estimating proportional contribution of breeding populations to non-breeding flocks. We also provide preliminary support for the inclusion of the two populations in the single subspecies C. a. alba, a conclusion based on morphology (Engelmoer and Roselaar 1998). However, assessment of the status of C. a. alba and the Nearctic subspecies C. a. rubidus still awaits an examination of global genetic population structure including individuals from all flyways.
Long-distance migratory shorebirds include species with deeply diverged subspecific lineages that apparently survived the LGM in distinct populations, such as Dunlin C. alpina (Buehler and Baker 2005; Wenink et al. 1993) and Black-tailed Godwit Limosa limosa (Höglund et al. 2009; Trimbos et al. 2014), and also those which presumably underwent bottlenecks in one or few LGM refugia and only recently expanded into multiple flyway populations, such as Red Knot (Buehler and Baker 2005; Buehler et al. 2006), Curlew Sandpiper C. ferruginea (Wennerberg and Burke 2001), and Ruddy Turnstone Arenaria interpres (Wenink et al. 1994). In mtDNA, the latter group are distinguishable by lower sequence variation and shallow divergences (i.e., star-like networks with haplotypes differing by only single base-pair changes). In this respect, Sanderlings and Red Knots appear similar: with comparable sample sizes in analogous populations (n = 12 C. c. canutus, 15 C. c. islandica), Buehler and Baker (2005) also detected nine variable sites accounting for 12 haplotypes in the control region (675 bp of CR I and III). When restricted to the 249-bp segment of the hypervariable CR I considered in both studies, there were just 5–6 variable sites in Sanderlings and Red Knots (compared to 30 variable sites in Dunlin; Wenink et al. 1993). Nucleotide diversity was also similarly low in the two species (0.0016–0.0033 in Greenland and Siberia).
Contrasting genetic patterns in currently co-distributed species may reflect different phylogeographic histories (Zink 1996). In Red Knots, Siberian and Greenlandic populations are clearly differentiated in mtDNA (F ST = 0.19), each containing a common private haplotype, and diverged into recognized subspecies approximately 23,000 years ago (CI 7000–59,000; Buehler and Baker 2005). In Sanderlings, analogous populations were much more weakly differentiated in mtDNA, featured no common private haplotypes, and appear to have diverged much more recently. Divergence time estimates are subject to uncertainty regarding mutation rates; we used the same mtDNA mutation rate as Buehler and Baker (2005) to enable direct comparison. Our T div estimates must be viewed as preliminary, as our IMa2 analyses failed to fully converge, probably due to the combination of weak differentiation and low sample size. However, independent parameter estimates from models for mtDNA and microsatellites consistently produced T div estimates of <10,000 years (Table 2). The comparatively recent T div estimates for microsatellite loci are consistent with an ongoing divergence with incompletely sorted lineages; such conditions are quicker to resolve in mtDNA because lineage sorting is faster with a lower N e (Zink and Barrowclough 2008). Our T div estimates gain credibility from the associated ancestral N e estimates, which fall within the expected order of magnitude, considering the current global population estimate of 600,000–700,000 Sanderlings (Wetlands International 2013).
Buehler et al. (2006) proposed that Red Knots survived the LGM in two Palearctic refugia, and then sequentially colonized Nearctic flyways in a counter-clockwise direction from eastern Siberia. Four pieces of information now argue against a similar scenario in Sanderlings: (1) much weaker or more recent isolation of Greenlandic and Siberian breeding populations (this study); (2) a lack of morphological differences between these populations (Engelmoer and Roselaar 1998); (3) a small breeding population on the intermediate archipelago of Svalbard (Norderhaug 1989; Strøm 2006), which is absent in Red Knots (Fig. 1); and (4) the absence of a present-day breeding population in the Russian Far East, analogous to C. c. rogersi in Red Knots. Together, these observations do not support a counter-clockwise colonization of the Nearctic from Siberia by Sanderlings. Therefore, the contemporary co-distribution of the species may be a recent rather than a historical phenomenon.
In addition to differences in phylogeographic history or dispersal propensity, smaller F ST in Sanderlings could potentially arise from differences in effective population size, because coalescence time increases with N e. In fact, our estimates of ancestral N ef for mtDNA (N e −2 = ca. 22,500) are higher than the pairwise ancestral N ef estimate for C. c. canutus and C. c. islandica in Red Knots (10,969; Buehler and Baker 2005). However, these imprecise estimates are on the same order of magnitude and have overlapping confidence intervals. In Red Knots, species-wide (six global populations pooled) haplotype diversity was consistent with a genetic bottleneck and subsequent population expansion (Tajima’s D = −1.98) during the last 31,000 years (Buehler and Baker 2005). In Sanderlings, we found a much weaker and non-significant signal in the same direction (D = −0.73). Current census populations are substantially smaller in Sanderlings (ca. 270,000 for Siberia/Greenland combined) than in Red Knots (850,000 for C. c. canutus/islandica combined; Wetlands International 2013). This further supports a less dramatic expansion in Sanderlings, and may indicate that contrasting population processes affecting these ecologically similar species continue in the present.
References
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57:289–300
BirdLife International (2013) Species factsheets: IUCN Red list for birds. BirdLife International, Cambridge, UK. http://www.birdlife.org/datazone/species. Accessed May 2014
Buehler DM, Baker AJ (2005) Population divergence times and historical demography in red knots and dunlins. Condor 107:497–513
Buehler DM, Baker AJ, Piersma T (2006) Reconstructing palaeoflyways of the late Pleistocene and early Holocene red knot Calidris canutus. Ardea 94:485–498
Carter K, Kempenaers B (2007) Eleven polymorphic microsatellite markers for paternity analysis in the pectoral sandpiper, Calidris melanotos. Molec Ecol Notes 7:658–660
Engelmoer M, Roselaar CS (1998) Geographic variation in waders. Kluwer, Dordrecht
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fridolfsson A-K, Ellegren H (1999) A simple and universal method for molecular sexing of non-ratite birds. J Avian Biol 30:116–121
Gilg O, Yoccoz NG (2010) Explaining bird migration. Science 327:276–277
Green G, Greenwood J (1978) Joint biological expedition to North East Greenland, 1974. Dundee University NE Greenland Expedition, Dundee
Gudmundsson GA, Lindström Å (1992) Spring migration of Sanderlings Calidris alba through SW Iceland: wherefrom and whereto? Ardea 80:315–326
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760
Höglund J, Johansson T, Beintema A, Schekkerman H (2009) Phylogeography of the Black-tailed Godwit Limosa limosa: substructuring revealed by mtDNA control region sequences. J Ornith 150:45–53
Langston R (2002) Sanderling Calidris alba. In: Wernham CV, Toms MP, Marchant JH, Clark JA, Siriwardena GM, Baillie SR (eds) The migration atlas: movements of the birds of Britain and Ireland. T. & A.D, Poyser, London, pp 297–299
Lappo EG, Tomkovich PS, Syroechkovski EE (2012) Atlas of breeding waders in the Russian Arctic. UF Ofsetnaya Pechat, Moscow
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lopes RJ, Alves JA, Gill JA, Gunnarsson TG, Hooijmeijer JC, Lourenço PM, Masero JA, Piersma T, Potts PM, Rabaçal B (2013) Do different subspecies of Black-tailed Godwit Limosa limosa overlap in Iberian wintering and staging areas? Validation with genetic markers. J Ornithol 154:35–40
Luttikhuizen PC, Bol A, Witte H, van Bleijswijk J, Haddrath O, Baker AJ, Piersma T, Reneerkens J (2011) Novel and cross-species microsatellite markers for parentage analysis in Sanderling Calidris alba. J Ornithol 152:807–810
Macwhirter B, Austin-Smith P, Jr., Kroodsma D (2002) Sanderling (Calidris alba). In: Poole A (ed) The birds of North America online. Cornell Lab of Ornithology, Ithaca, NY, USA. http://bna.birds.cornell.edu.bnaproxy.birds.cornell.edu/bna/species/653. Accessed Feb 2014
Meltofte H, Rasch M (2008) The study area at Zackenberg. In: Meltofte H, Christensen T, Elberling B, Forchhammer M, Rasch M (eds) Ten years of monitoring and research at Zackenberg Research Station, Northeast Greenland. Advances in ecological research, vol 40. Academic Press, San Diego, pp 101–110
Meltofte H, Blew J, Frikke J, Rösner H-U, Smit CJ (1994) Numbers and distribution of waterbirds in the Wadden Sea: results and evaluation of 36 simultaneous counts in the Dutch-German-Danish Wadden Sea 1980–1991. International Waterfowl and Wetlands Research Bureau Publication 34 & Wader Study Group Bulletin, vol 74 special issue
Neff BD, Gross MR (2001) Microsatellite evolution in vertebrates: inference from AC dinucleotide repeats. Evolution 55:1717–1733
Norderhaug M (1989) Svalbards Fugler. vol (In Norwegian). Dreyer, Oslo
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Reneerkens J, Morrison RIG, Coulomb G (2008) First re-sighting in Europe of an individually marked Sanderling Calidris alba from Ellesmere Island. Wader Study Group Bull 115:116–118
Reneerkens J, Benhoussa A, Boland H, Collier M, Grond K, Günther K, Hallgrimsson GT, Hansen J, Meissner W, de Meulenaer B, Ntiamoa-Baidu Y, Piersma T, Poot M, van Roomen M, Summers RW, Tomkovich PS, Underhill LG (2009) Sanderlings using African-Eurasian flyways: a review of current knowledge. Wader Study Group Bull 116:2–20
Reneerkens J, van Veelen P, van der Velde M, Luttikhuizen P, Piersma T (2014) Within-population variation in mating system and parental care patterns in the Sanderling (Calidris alba) in northeast Greenland. Auk 131:235–247
Scott D (2009) Sanderling. In: Delany S, Scott D, Dodman T, Stroud D (eds) An atlas of wader populations in African and western Eurasia. Wetlands International, Wageningen, The Netherlands
Soloviev MY, Tomkovich PS (1995) Biometrics of Sanderlings Calidris alba from the Taimyr. Ring Migr 16:91–99
Strøm H (2006) Sanderling Calidris alba. In: Kovacs KM, Lydersen C, Polarinstitutt N (eds) Birds and mammals of Svalbard. Norwegian Polarinstitute, Tromsø
Stroud DA, Davidson NC, West R, Scott DA, Haanstra L, Thorup O, Ganter B, Delaney S (2004) Status of migratory wader populations in Africa and western Eurasia in the 1990s, vol 15. International Wader Studies, International Wader Study Group
Tomkovich PS, Serra L (1999) Morphometrics and prediction of breeding origin in some Holarctic waders. Ardea 87:289–300
Trimbos KB, Doorenweerd C, Kraaijeveld K, Musters C, Groen NM, de Knijff P, Piersma T, de Snoo GR (2014) Patterns in nuclear and mitochondrial DNA reveal historical and recent isolation in the Black-tailed Godwit (Limosa limosa). PLoS One 9:e83949
Tulp I, Schekkerman H, Piersma T, Jukema J, de Goeij P, van de Kam J (1998) Breeding waders at Cape Sterlegova, Northern Taimyr, in 1994. WIWO report 61. Stichting Werkgroep Internationaal Watervogelsonderzoek (WIWO), report 61, Zeist
Underhill LG, Tree A, Oschadleus H, Parker V (1999) Review of ring recoveries of waterbirds in southern Africa. University of Cape Town, South Africa, Avian Demography Unit
Wenink PW, Baker AJ, Tilanus MG (1993) Hypervariable-control-region sequences reveal global population structuring in a long-distance migrant shorebird, the Dunlin (Calidris alpina). Proc Natl Acad Sci 90:94–98
Wenink PW, Baker AJ, Tilanus M (1994) Mitochondrial control-region sequences in two shorebird species, the Turnstone and the Dunlin, and their utility in population genetic studies. Molec Biol Evol 11:22–31
Wennerberg L, Burke T (2001) Low genetic differentiation between Curlew Sandpiper (Calidris ferruginea) populations with highly divergent migratory directions shown by mitochondrial DNA and microsatellite analysis. In: Wennerberg, L. Genetic variation and migration in waders. Ph.D. dissertation, Department of Ecology, Lund University
Werle E, Schneider C, Renner M, Völker M, Fiehn W (1994) Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res 22:4354–4355
Wetlands International (2013) Waterbird population estimates, 5th edn. Wetlands International, The Netherlands. http://wpe.wetlands.org/. Accessed May 2013
Whittaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM (2003) Likelihood-based estimation of microsatellite mutation rates. Genetics 164:781–787
Zink RM (1996) Comparative phylogeography in North American birds. Evolution 50:308–317
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Molec Ecol 17:2107–2121
Acknowledgments
We thank M. van der Velde, P. van Veelen, and M. Bérubé for technical assistance. We thank M. Peck at the Royal Ontario Museum for making Siberian samples available. Fieldwork by TP and the team at Sterlegova in 1994 was made possible by 80 private benefactors and small grants from the Netherlands Organisation for Scientific Research (NWO) and the Ministry of Agriculture, Nature Management and Fisheries. Fieldwork by P.S.T. at Knipovich Bay was conducted in a field camp of the International Arctic Expedition of the Institute of Evolutionary Morphology and Animal Ecology, Russian Academy of Sciences, with financial and logistical support of the late Prof. E.E. Syroechkovskiy. Fieldwork by J.R. in Greenland was supported by INTERACT (Grant Agreement No. 262693) under the European Community’s Seventh Framework Programme, and facilitated by the Zackenberg Logistics team at the Department of Bioscience, Aarhus University, Roskilde. J.R. was supported by Waddenfonds through the Metawad project (WF209955, awarded to T.P.). P.S.T. was supported by Grant No. 14-50-00029 of the Russian Science Fund. T.P. was supported by BirdLife Netherlands and the World Wide Fund for Nature, Netherlands, through the Chair in Global Flyway Ecology, University of Groningen. We thank R. Zink for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wink.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Conklin, J.R., Reneerkens, J., Verkuil, Y.I. et al. Low genetic differentiation between Greenlandic and Siberian Sanderling populations implies a different phylogeographic history than found in Red Knots. J Ornithol 157, 325–332 (2016). https://doi.org/10.1007/s10336-015-1284-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-015-1284-4