Abstract
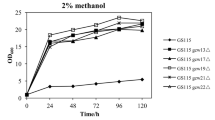
Glycosylphosphatidylinositol (GPI)-anchored glycoproteins have diverse intrinsic functions in yeasts, and they also have different uses in vitro. The GPI-modified cell wall proteins GCW21, GCW51, and GCW61 of Pichia pastoris were chosen as anchoring proteins to construct co-expression strains in P. pastoris GS115. The hydrolytic activity and the amount of Candida antarctica lipase B (CALB) displayed on cell surface increased significantly following optimization of the fusion gene dosage and combination of the homogeneous or heterogeneous cell wall proteins. Maximum CALB hydrolytic activity was achieved at 4920 U/g dry cell weight in strain GS115/CALB-GCW (51 + 51 + 61 + 61) after 120 h of methanol induction. Changes in structural morphology and the properties of the cell surfaces caused by co-expression of fusion proteins were observed by transmission electron microscopy (TEM) and on plates containing cell-wall-destabilizing reagent. Our results suggested that both the outer and inner cell layers were significantly altered by overexpression of GPI-modified cell wall proteins. Interestingly, quantitative analysis of the inner layer components showed an increase in β-1,3-glucan, but no obvious changes in chitin in the strains overexpressing GPI-modified cell wall proteins.





Similar content being viewed by others
References
Bulawa CE (1992) CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol 12(4):1764–1776. doi:10.1128/MCB.12.4.1764
Bulawa CE, Slater M, Cabib E, Au-Young J, Sburlati A, Adair WL Jr, Robbins PW (1986) The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell 46(2):213–225. doi:10.1016/0092-8674(86)90738-5
Carotti C, Ferrario L, Roncero C, Valdivieso MH, Duran A, Popolo L (2002) Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast 19(13):1113–1124. doi:10.1002/yea.905
Cos O, Serrano A, Montesinos JL, Ferrer P, Cregg JM, Valero F (2005) Combined effect of the methanol utilization (Mut) phenotype and gene dosage on recombinant protein production in Pichia pastoris fed-batch cultures. J Biotechnol 116(4):321–335. doi:10.1016/j.jbiotec.2004.12.010
Fukuda H, Hama S, Tamalampudi S, Noda H (2008) Whole-cell biocatalysts for biodiesel fuel production. Trends Biotechnol 26(12):668–673. doi:10.1016/j.tibtech.2008.08.001
Guo D, Xu Y, Kang Y, Han S, Zheng S (2016) Synthesis of octyl-β-d-glucopyranoside catalyzed by Thai rosewood β-glucosidase-displaying Pichia pastoris in an aqueous/organic two-phase system. Enzyme Microb Tech 85:90–97. doi:10.1016/j.enzmictec.2015.07.006
Guo DH, Zi J, Xu YS, Ping W, Ying L, Han SY, Zheng SP (2015) Scaling-up the synthesis of myristate glucose ester catalyzed by a CALB-displaying Pichia pastoris whole-cell biocatalyst. Enzyme Microb Tech 75–76:30–36. doi:10.1016/j.enzmictec.2015.04.002
He X, Liu N, Li W, Zhang Z, Zhang B, Ma Y (2008) Inducible and constitutive expression of a novel thermostable alkaline β-mannanase from alkaliphilic Bacillus sp. N16-5 in Pichia pastoris and characterization of the recombinant enzyme. Enzyme Microb Tech 43 (1):13–18. doi:10.1016/j.enzmictec.2008.03.011
Imai K, Noda Y, Adachi H, Yoda K (2005) A novel endoplasmic reticulum membrane protein Rcr1 regulates chitin deposition in the cell wall of Saccharomyces cerevisiae. J Biol Chem 280(9):8275–8284. doi:10.1074/jbc.M409428200
Jin Z, Han SY, Zhang L, Zheng SP, Wang Y, Lin Y (2013) Combined utilization of lipase-displaying Pichia pastoris whole-cell biocatalysts to improve biodiesel production in co-solvent media. Bioresour Technol 130(2):102–109. doi:10.1016/j.biortech.2012.12.020
Kapteyn JC, Montijn RC, Vink E, de la Cruz J, Llobell A, Douwes JE, Shimoi H, Lipke PN, Klis FM (1996) Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked beta-1,3-/beta-1,6-glucan heteropolymer. Glycobiology 6(3):337–345. doi:10.1093/glycob/6.3.337
Kollar RR, Reinhold BB, Petráková EE, Yeh HJC, Ashwell GG, Drgonová JJ, Kapteijn JC, Klis FM, Cabib EE (1997) Architecture of the yeast cell wall. b (1-6)-glucan interconnects mannoprotein, b (1-3)-glucan, and chitin. J Biol Chem 272(28):17762–17775. doi:10.1074/jbc.272.28.17762
Kondo A, Ueda M (2004) Yeast cell-surface display-applications of molecular display. Appl Microbiol Biot 64(1):28–40. doi:10.1007/s00253-003-1492-3
Kordel M, Hofmann B, Schomburg D, Schmid RD (1991) Extracellular lipase of Pseudomonas sp. strain ATCC 21808: purification, characterization, crystallization, and preliminary X-ray diffraction data. J Bacteriol 173(15):4836–4841
Kuroda K, Ueda M (2013) Arming technology in yeast-novel strategy for whole-cell biocatalyst and protein engineering. Biomolecules 3(3):632–650. doi:10.3390/biom3030632
Lee SY, Choi J, Xu Z (2003) Microbial cell-surface display. Trends Biotechnol 21:45–52. doi:10.1016/S0167-7799(02)00006-9
Li C, Lin Y, Huang Y, Liu X, Liang S (2014) Citrobacter amalonaticus phytase on the cell surface of Pichia pastoris exhibits high pH stability as a promising potential feed supplement. PLoS One 9(12):e114728. doi:10.1371/journal.pone.0114728
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)). Methods 25(4):402–408. doi:10.1006/meth.2001.1262
Lu CF, Kurjan J, Lipke PN (1994) A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol Cell Biol 14(7):4825–4833. doi:10.1128/MCB.14.7.4825
Lu CF, Montijn RC, Brown JL, Klis F, Kurjan J, Bussey H, Lipke PN (1995) Glycosyl phosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta 1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol 128(3):333–340
Martin H, Dagkessamanskaia A, Satchanska G, Dallies N, Francois J (1999) KNR4, a suppressor of Saccharomyces cerevisiae cwh mutants, is involved in the transcriptional control of chitin synthase genes. Microbiology 145 (Pt1) (1):249–258. doi:10.1099/13500872-145-1-249
Pittet M, Conzelmann A (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Et Biophys Acta 1771(1771):405–420. doi:10.1016/j.bbalip.2006.05.015
Ram AF, Kapteyn JC, Montijn RC, Caro LH, Douwes JE, Baginsky W, Mazur P, van den Ende H, Klis FM (1998) Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol 180(6):1418–1424
Reissig JL, Storminger JL, Leloir LF (1955) A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem 217(2):959–966
Roman K, Reinhold BB, Gilbert A (1997) Architecture of the yeast cell wall. J Biol Chem 272(3):17762–17775
Sato N, Matsumoto T, Ueda M, Tanaka A, Fukuda H, Kondo A (2002) Long anchor using Flo1 protein enhances reactivity of cell surface-displayed glucoamylase to polymer substrates. Appl Microbiol Biot 60(4):469–474. doi:10.1007/s00253-002-1121-6
Sekiya-Kawasaki M, Abe M, Saka A, Watanabe D, Kono K, Minemura-Asakawa M, Ishihara S, Watanabe T, Ohya Y (2002) Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, Saccharomyces cerevisiae. Genetics 162(2):663–676. doi:10.3103/S0096392510040279
Sha C, Yu XW, Li F, Xu Y (2013) Impact of gene dosage on the production of lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris. Appl Biochem Biotechnol 169(4):1160–1172. doi:10.1007/s12010-012-0050-9
Su GD, Huang DF, Han SY, Zheng SP, Lin Y (2010) Display of Candida antarctica lipase B on Pichia pastoris and its application to flavor ester synthesis. Appl Microbiol Biot 86(5):1493–1501. doi:10.1007/s00253-009-2382-0
Sun YF, Lin Y, Zhang JH, Zheng SP, Ye YR, Liang XX, Han SY (2012) Double Candida antarctica lipase B co-display on Pichia pastoris cell surface based on a self-processing foot-and-mouth disease virus 2A peptide. Appl Microbiol Biot 96(6):1539–1550. doi:10.1007/s00253-012-4264-0
Fukuda TT, Kondo HA (2006) Construction of a Pichia pastoris cell-surface display system using Flo1p anchor system. Biotechnol Prog 22(4):989–993. doi:10.1021/bp060133
Tanino T, Ohno T, Aoki T, Fukuda H, Kondo A (2007) Development of yeast cells displaying Candida antarctica lipase B and their application to ester synthesis reaction. Appl Microbiol Biotechnol 75(6):1319–1325. doi:10.1007/s00253-007-0959-z
Van der Vaart JM, te Biesebeke R, Chapman JW, Klis FM, Verrips CT (1996) The beta-1, 6-glucan containing side-chain of cell wall proteins of Saccharomyces cerevisiae is bound to the glycan core of the GPI moiety. FEMS Microbiol Lett 145(3):401–407. doi:10.1016/S0378-1097(96)00440-5
Wang P, He J, Sun Y, Reynolds M, Zhang L, Han S, Liang S, Sui H, Lin Y (2016) Display of fungal hydrophobin on the Pichia pastoris cell surface and its influence on Candida antarctica lipase B. Appl Microbiol Biotechnol 100(13):1–13. doi:10.1007/s00253-016-7431-x
Yan J, Zheng X, Li S (2014) A novel and robust recombinant Pichia pastoris yeast whole cell biocatalyst with intracellular overexpression of a Thermomyces lanuginosus lipase: preparation, characterization and application in biodiesel production. Bioresour Technol 151(1):43–48. doi:10.1016/j.biortech.2013.10.037
Yin QY, de Groot PW, Dekker HL, De JL, Klis FM, de Koster CG (2005) Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J Biol Chem 280(21):20894–20901. doi:10.1074/jbc.M500334200
Zhang L, Liang SL, Zhou XY, Jin Z, Jiang FC, Han SY, Zheng SP, Lin Y (2013) Screening for glycosylphosphatidylinositol-modified cell wall proteins in Pichia pastoris and their recombinant expression on the cell surface. Appl Environ Microb 79(18):5519–5526. doi:10.1128/AEM.00824-13
Acknowledgements
The authors thank Jie He, Shufeng Sun, and Matthew Reynolds at the Wadsworth Center for their assistance in yeast cell growth, specimen preparation, and electron microscopy. This work was supported in whole by the grants to Ying Lin from, the China National High Technology Research and Development Program (863-2012AA022205), High Technology Research and Development of Guangdong Province (2012A080800013), and The Recruitment Program of Leading Talents in Innovation and Entrepreneurship of Guangzhou (LCY201322), and in part by the National Institute of Health (NIH) Grants GM097010 and GM101026 to Haixin Sui.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported in whole by the grants to Ying Lin from, the China National High Technology Research and Development Program (863-2012AA022205), High Technology Research and Development of Guangdong Province (2012A080800013), and The Recruitment Program of Leading Talents in Innovation and Entrepreneurship of Guangzhou (LCY201322), and in part by the National Institute of Health (NIH) Grants GM097010 and GM101026 to Haixin Sui.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any of the authors.
Rights and permissions
About this article
Cite this article
Wang, P., Zhang, L., Fisher, R. et al. Accurate analysis of fusion expression of Pichia pastoris glycosylphosphatidylinositol-modified cell wall proteins. J Ind Microbiol Biotechnol 44, 1355–1365 (2017). https://doi.org/10.1007/s10295-017-1962-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1962-8