Abstract
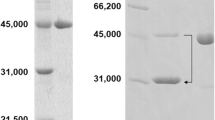
α-Amino-ε-caprolactam (ACL) racemizing activity was detected in a putative dialkylglycine decarboxylase (EC 4.1.1.64) from Citreicella sp. SE45. The encoding gene of the enzyme was cloned and transformed in Escherichia coli BL21 (DE3). The molecular mass of the enzyme was shown to be 47.4 kDa on SDS–polyacrylamide gel electrophoresis. The enzymatic properties including pH and thermal optimum and stabilities were determined. This enzyme acted on a broad range of amino acid amides, particularly unbranched amino acid amides including l-alanine amide and l-serine amide with a specific activity of 17.5 and 21.6 U/mg, respectively. The Km and Vmax values for d- and l-ACL were 5.3 and 2.17 mM, and 769 and 558 μmol/min.mg protein, respectively. Moreover, the turn over number (Kcat) and catalytic efficiency (Kcat/Km) of purified ACL racemase from Citreicella sp. SE45 using l-ACL as a substrate were 465 S−1 and 214 S−1mM−1, respectively. The new ACL racemase from Citreicella sp. SE45 has a potential to be used as the biocatalytic application.

Similar content being viewed by others
References
Ahmed SA, Esaki N, Soda K (1982) Purification and properties of α-amino-ε-caprolactam racemase from Achromobacter obae. FEBS Lett 150:370–374. doi:10.1016/0014-5793(82)80770-9
Ahmed SA, Esaki N, Tanaka H, Soda K (1983) Properties of α-amino-ε-caprolactam racemase from Achromobacter obae. Agric Biol Chem 47:1881–1893. doi:10.1080/00021369.1983.10865864
Asano Y (2010) Tools for enzyme discovery. In: Baltz RH, Davies JE, Demain AL (eds) Manual of industrial microbiology and biotechnology. ASM Press, Washington DC, pp 441–452
Asano Y, Lübbehüsen TL (2000) Enzymes acting on peptides containing d-amino acid. J Biosci Bioeng 89:295–306. doi:10.1016/S1389-1723(00)88949-5
Asano Y, Yamaguchi S (2005) Discovery of amino acid amides as new substrates for α-amino-ε-caprolactam racemase from Achromobacter obae. J Mol Catal B Enzym 36:22–29. doi:10.1016/j.molcatb.2005.07.003
Asano Y, Yamaguchi S (2005) Dynamic kinetic resolution of amino acid amide catalyzed by d-aminopeptidase and α-amino-ε-caprolactam racemase. J Am Chem Soc 127:7696–7697. doi:10.1021/ja050300m
Baek DH, Kwon SJ, Hong SP, Kwak MS, Lee MH, Song JJ, Lee SG, Yoon KH, Sung MH (2003) Characterization of a thermostable d-stereospecific alanine amidase from Brevibacillus borstelensis BCS-1. Appl Environ Microbiol 69:980–986. doi:10.1128/AEM.69.2.980-986.2003
Baek DH, Song JJ, Lee SG, Kwon SJ, Asano Y, Sung MH (2003) New thermostable d-methionine amidase from Brevibacillus borstelensis BCS-1 and its application for d-phenylalanine production. Enzyme Microb Tech 32:131–139. doi:10.1016/S0141-0229(02)00268-5
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Buchan A, Neidle EL, Moran MA (2001) Diversity of the ring-cleaving dioxygenase genepcaH salt marsh bacterial community. Appl Environ Microbiol 67:5801–5809. doi:10.1128/AEM.67.12.5801-5809.2001
Fukumura T (1976) Hydrolysis of l-α-amino-ɛ-caprolactam by yeasts. J Agric Biol Chem 40:1695–1698. doi:10.1080/00021369.1976.10862305
Fukumura T (1976) Screening, classification and distribution of l-α-amino-ɛ-caprolactam-hydrolyzing yeasts. J Agric Biol Chem 40:1687–1689. doi:10.1080/00021369.1976.10862304
Fukumura T (1977) Bacterial racemization of α-amino-ε-caprolactam. Agric Biol Chem 41:1321–1325. doi:10.1080/00021369.1977.10862695
Fukumura T (1977) Conversion of d- and dl-α-amino-ε-caprolactam into l-lysine using both yeast cells and bacterial cells. Agric Biol Chem 41:1327–1330. doi:10.1080/00021369.1977.10862696
Hendrick ME (1991) Alitame. Food Sci Technol 48:29–38
Hermes HFM, Sonke T, Peters PJH, van Balken JAM, Kamphuis J, Dijkhuizen L, Meijer EM (1993) Purification and characterization of an l-aminopeptidase from Pseudomonas putida ATCC 12633. Appl Environ Microbiol 59:4330–4334
Hermes HFM, Tandler RF, Sonke T, Dijkhuizen L, Meijer EM (1994) Purification and characterization of an l-amino amidase from Mycobacterium neoaurum ATCC 25795. Appl Environ Microbiol 60:153–159
Hohenester E, Keller JW, Jansonius JN (1994) An alkali metal ion size-dependent switch in the active site structure of dialkylgycine decarboxylase. Biochemistry 46:13561–13570
Hongpattarakere T, Komeda H, Asano Y (2005) Purification, characterization, gene cloning and nucleotide sequencing of d-stereospecific amino acid amidase from soil bacterium: Delftia acidovorans. J Ind Microbiol Biotechnol 32:567–576. doi:10.1007/s10295-005-0246-x
Komeda H, Asano Y (2000) Gene cloning, nucleotide sequencing, and purification and characterization of the d-stereospecific amino acid amidase from Ochrobactrum anthropi SV3. J Biochem 267:2028–2035. doi:10.1046/j.1432-1327.2000.01208.x
Komeda H, Asano Y (2008) A novel d-stereoselective amino acid amidase from Brevibacterium iodinum: gene cloning, expression and characterization. Enzyme Microb Tech 43:276–283. doi:10.1016/j.enzmictec.2008.03.008
Komeda H, Harada H, Washika S, Sakamoto T, Ueda M, Asano Y (2004) S-Stereoselective piperazine-2-tert-butylcarboxamide hydrolase from Pseudomonas azotoformans IAM 1603 is a novel l-amino acid amidase. Eur J Biochem 271:1465–1475. doi:10.1111/j.1432-1033.2004.04056.x
Komeda H, Hariyama N, Asano Y (2006) l-Stereoselective amino acid amidase with broad substrate specificity from Brevundimonas diminuta: characterization of a new member of the leucine aminopeptidase family. Appl Microbiol Biotechnol 70:412–421. doi:10.1007/s00253-005-0068-9
Krieg L, Ansorge-Schumacher MB, Kula MR (2002) Screening for amidases: isolation and characterization of a novel d-amidase from Variovorax paradoxus. Adv Synth Catal 344:965–973. doi:10.1002/1615-4169(200210)344:9<965:AID-ADSC965>3.0.CO;2-Z
Laemmli U (1970) Cleavage of structural proteins during the head of bacteriophage T4. Nature 277:860–865. doi:10.1038/227680a0
Lim YH, Yokoigawa K, Esaki N, Soda K (1993) A new amino acid racemase with threonine α-epimerase activity from Pseudomonas putida: purification and characterization. J Bacteriol 175:4213–4217
Liu W, Peterson PE, Langston JA, Jin X, Zhou X, Fisher AJ, Toney MD (2005) Kinetic and crystallographic analysis of active site mutants of Escherichia coli gamma-aminobutyrate aminotransferase. Biochemistry 44:2982–2992. doi:10.1021/bi048657a
Matsumura S (2013) Study on substrate specificity of amino acid amide racemase from Achromobacter obae. M.Sc. Thesis. Toyama Prefectural University, Japan
McNicholas S, Potterton E, Wilson KS, Noble EM (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr Sect D Biol Crystallogr 67:386–394. doi:10.1107/S0907444911007281
Okazaki S, Suzuki A, Mizushima T, Kawano T, Komeda T, Asano Y, Yamane T (2009) The novel structure of a Pyridoxal 5′-phosphate-dependent fold-type I racemase, α-amino-ε-caprolactam racemase from Achromobacter obae. Biochemistry 48:941–950. doi:10.1021/bi801574p
Payoungkiattikun W, Okazaki S, Nakano S, Ina A, H-Kittikun A, Asano Y (2015) In silico identification for α-amino-ε-caprolactam racemases by using information on the structure and function relationship. Appl Biochem Biotech 176:1303–1314. doi:10.1007/s12010-015-1647-6
Pellegata R, Pinza M, Pifferi G (1978) An improved synthesis of γ-, δ-, and ε-lactams. Synthesis 8:614–616. doi:10.1055/s-1978-24834
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. Cold Spring Harbor Laboratory Press, New York
Singh M, Yadav A, Ma X, Amoah E (2010) Plasmid DNA transformation in Escherichia coli: effect of heat shock temperature, duration, and cold incubation of CaCl2 treated cells. Int J Biochem Biotechnol 6:561–568
Sonke T (2008) Novel development in the chemo-enzymatic synthesis of enantiopure α-hydrogen and α, α-disubstituted α-amino acids and derivative. PhD. Thesis. The University of Amsterdam, Netherland
Steffen-Munsberg F, Vickers C, Kohls H, Land H, Mallin H, Nobili A, Skalden L, van den Bergh T et al (2015) Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol Adv 33:566–604. doi:10.1016/j.biotechadv.2014.12.012
van den Tweel WJJ, van Dooren TJGM, de Jonge PH, Kaptein B, Duchateau ALL, Kamphuis J (1993) Ochrobactrum anthropi NCIMB 40321: a new biocatalyst with broad-spectrum l-specific amidase activity. Appl Microbiol Biotechnol 39:296–300. doi:10.1007/BF00192081
Wakayama M, Moriguchi M (2001) Comparative biochemistry of bacterial N-acyl-d-amino acid amidohydrolase. J Mol Catal B Enzym 12:15–25. doi:10.1016/S1381-1177(00)00199-5
Yamada H, Shimizu S (1988) Microbial and enzymatic processes for the production of biologically and chemically useful compounds. Angew Chem Int Ed Engl 27:622–642. doi:10.1002/anie.198806221
Yamaguchi S, Komeda H, Asano Y (2007) New enzymatic method of chiral amino acid synthesis by dynamic kinetic resolution of amino acid amides: use of stereoselective amino acid amidases in the presence of α-amino-ε-caprolactam racemase. Appl Environ Microbiol 73:5370–5373. doi:10.1128/AEM.00807-07
Zhou X, Kay S, Toney MD (1998) Coexisting kinetically distinguishable forms of dialkylglycine decarboxylase engendered by alkali metal ions. Biochemistry 37:5761–5769
Acknowledgments
This work was financially supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/099/2551) and the Japan Student Services Organization (JASSO). This research was partly supported by Grant-in-Aids for Scientific Research (A) (23248015) and (B) (26292041) to Y. Asano from the Japan Society for the Promotion of Science (JSPS). This work was also partly supported by the Exploratory Research for Advanced Technology (ERATO) Asano Active Enzyme Molecule Project of Japan Science and Technology Agency (JST). The author greatly appreciates Asst. Prof. Ken-ichi Fuhshuku for the preparation of chiral ACLs and enantiomerically pure amino acid amides. This work was performed at Toyama Prefectural University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no completing interests.
Ethical approval
This study does not contain any experiment with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Payoungkiattikun, W., Okazaki, S., Ina, A. et al. Characterization of an α-amino-ɛ-caprolactam racemase with broad substrate specificity from Citreicella sp. SE45. J Ind Microbiol Biotechnol 44, 677–685 (2017). https://doi.org/10.1007/s10295-016-1825-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1825-8