Abstract
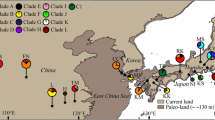
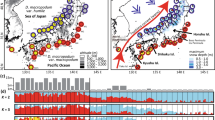
A phylogeographic study of four tree species (Padus grayana, Euonymus oxyphyllus, Magnolia hypoleuca, and Carpinus laxiflora) growing in Japanese deciduous broad-leaved forests was conducted based on chloroplast DNA (cpDNA) variations. Using nucleotide sequences of 702–1,059 bp of intergenic spacers of cpDNA, 20, 27, eight, and eight haplotypes were detected among 251, 251, 226, and 262 individuals sampled from 67, 79, 75, and 71 populations of the above species, respectively. The geographical pattern of the cpDNA variations was highly structured in each species, and the following three regional populations were genetically highly differentiated among all four species: (1) the Sea of Japan-side area, (2) the Kanto region, and (3) southwestern Japan. Based on some interspecific similarities among the phylogeographic patterns, the following migration scenario of Japanese deciduous broad-leaved forests was postulated. During the last glacial maximum (LGM), the forests were separately distributed in six regions. After LGM, as the climate warmed, the forests in eastern Japan separately expanded from each of the refugia along the Sea of Japan-side or along the Pacific Ocean-side. In contrast, those in southwestern Japan retreated and moved to high altitudes from each of the continuous forests.






Similar content being viewed by others
References
Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J (2000) Molecular analysis of plant migration and refugia in the Arctic. Science 289:1343–1346
Aoki K, Suzuki T, Hu T, Murakami N (2004) Phylogeography of the component species of broad-leaved evergreen forests in Japan, based on chloroplast DNA variation. J Plant Res 117:77–94
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Bivand R (2009) The spdep package. Comprehensive R archive network, version 0.4-34
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Corriveau JL, Coleman AW (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot 75:1443–1458
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinforma Online 1:47–50
Fujii N, Senni K (2006) Phylogeography of Japanese alpine plants: biogeographic importance of alpine region of Central Honshu in Japan. Taxon 55:43–52
Fujii N, Ueda K, Watano Y, Shimizu T (1997) Intraspecific sequence variation of chloroplast DNA in Pedicularis chamissonis Steven (Scrophulariaceae) and geographic structuring of the Japanese “Alpine” plants. J Plant Res 110:195–207
Fujii N, Tomaru N, Okuyama K, Koike T, Mikami T, Ueda K (2002) Chloroplast DNA phylogeography of Fagus crenata (Fagaceae) in Japan. Plant Syst Evol 232:21–33
Godbout J, Jaramillo-Correa JP, Beaulieu J, Bousquet J (2005) A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range North American conifer. Mol Ecol 141:3497–3512
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamilton MB (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8:521–523
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources. Sinauer, Sunderland, pp 43–63
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124
Heuertz M, Fineschi S, Anzidei M, Pastorelli R, Salvini D, Paule L, Frascaria-Lacoste N, Hardy OJ, Vekemans X, Vendramin GG (2004) Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol Ecol 13:3437–3452
Hewitt GM (1996) Some genetic consequences of ice ages, and their role, in divergence and speciation. Biol J Linn Soc Lond 58:247–276
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B 359:183–195
Hiraoka K, Tomaru N (2009) Genetic divergence in nuclear genomes between populations of Fagus crenata along the Japan Sea and Pacific sides of Japan. J Plant Res 122:269–282
Ikeda H, Setoguchi H (2007) Phylogeography and refugia of the Japanese endemic alpine plant, Phyllodoce nipponica Makino (Ericaceae). J Biogeogr 34:169–176
Ingvarsson PK, Ribstein S, Taylor DR (2003) Molecular evolution of insertions and deletions in the chloroplast DNA genome of silence. Mol Biol Evol 20:1737–1740
Iwasaki T, Aoki K, Seo A, Murakami N (2006) Intraspecific sequence variation of chloroplast DNA among the component species of deciduous broad-leaved forests in Japan. J Plant Res 119:539–552
Kamei T, Research Group for the Biogeography from Würm Glacial (1981) Fauna and flora of the Japanese islands in the last glacial time. Quaternary Res 20:191–205 (in Japanese with English abstract)
Kanno M, Yokoyama J, Suyama Y, Ohyama M, Itoh T, Suzuki M (2004) Geographical distribution of two haplotypes of chloroplast DNA in four oak species (Quercus) in Japan. J Plant Res 117:311–317
Kira T (1977) A climatological interpretation of Japanese vegetation zones. In: Miyawaki A, Tüxen R (eds) Vegetation science and environmental protection. Maruzen Co, Ltd, Tokyo, pp 21–30
Kira T (1991) Forest ecosystems of east and southeast Asia in a global perspective. Ecol Res 6:185–200
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latałowa M, Litt T, Paule T, Roure JM, Tantau I, Knaap WO, Petit RJ, Beaulieu J (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–221
McCauley DE (1995) The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol Evol 10:198–202
McCune B, Mefford MJ (1999) PC-ORD. Multivariate analysis of ecological communities. MjM Software Design, Gleneden Beach, OR, USA
McCune B, Grace JB, Urban DL (2002) Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR, USA
McLachlan JS, Clark JS (2004) Reconstructing historical ranges with fossil data at continental scales. For Ecol Manag 197:139–147
Moran PAP (1950) Notes on continuous stochastic phenomena. Biometrika 37:17–23
Murata G (1972) Taxonomical notes 10. Acta Phytotax Geobot 25:45–53 (in Japanese)
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Nishizawa T, Watano Y (2000) Primer pairs suitable for PCR-SSCP analysis of chloroplast DNA in angiosperms. J Phytogeogr Taxon 48:63–66
Ohi T, Wakabayashi M, Wu S, Murata J (2003) Phylogeography of Stachyurus praecox (Stachyuraceae) in the Japanese archipelago based on chloroplast DNA haplotypes. J Jpn Bot 78:1–14
Ohta Y, Yonekura N (1987) Coastal line. In: Japan Association for Quaternary Research (ed) Quaternary maps of Japan. University of Tokyo Press, Tokyo, pp 70–72 (in Japanese)
Okaura T, Harada K (2002) Phylogeographical structure revealed by chloroplast DNA variation in Japanese beech (Fagus crenata Blume). Heredity 88:322–329
Okaura T, Quang ND, Ubukata M, Harada K (2007) Phylogeographic structure and late Quaternary population history of the Japanese oak Quercus mongolica var. crispula and related species revealed by chloroplast DNA variation. Genes Genet Syst 82:465–477
Ono Y (1984) Late glacial paleoclimate reconstructed from glacial and periglacial landforms in Japan. Geogr Rev Jpn Ser B 57:80–100 (in Japanese)
Peakall R, Smouse PE (2006) GENEALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petit RJ, Aguinagalde I, Beaulieu J, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Muller-Starck G, Demesure-Musch B, Palme A, Martin JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing. http://www.P-project.org/
Setoguchi H, Ohba H (1995) Phylogenetic relationships in Crossostylis (Rhizophoraceae) inferred from restriction site variation of chloroplast DNA. J Plant Res 108:87–92
Simmons MP, Ochoterena H (2000) Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49:369–381
Sneath PHA, Sokal PR (1973) Numerical taxonomy: the principles and practice of numerical classification. Freeman WH, San Francisco
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Takahara H, Sugita S, Harrison SP, Miyoshi N, Morita Y, Uchiyama T (2000) Pollen-based reconstructions of Japanese biomes at 0, 6000 and 18,000 14C yr BP. J Biogeogr 27:665–683
Takahashi T, Tani N, Taira H, Tsumura Y (2005) Microsatellite markers reveal high allelic variation in natural populations of Cryptomeria japonica near refugial areas of the last glacial period. J Plant Res 118:83–90
Terachi T (1993) Structural alterations of chloroplast genome and their significance to the higher plant evolution. Bull Inst Natl Land Util Dev Kyoto Sangyo Univ 14:138–148 (in Japanese with English summary)
Tomaru N, Mitsutsuji T, Takahashi M, Tsumura Y, Uchida K, Ohba K (1997) Genetic diversity in Fagus crenata (Japanese beech): influence of the distribution shift during the late-Quaternary. Heredity 78:241–251
Tomaru N, Takahashi M, Tsumura Y, Takahashi M, Ohba K (1998) Intraspecific variation and phylogeographic patterns of Fagus crenata (Fagaceae) mitochondrial DNA. Am J Bot 85:629–636
Tremblay NO, Schoen DJ (1999) Molecular phylogeography of Dryas integrifolia: glacial refugia and postglacial recolonization. Mol Ecol 8:1187–1198
Tsukada M (1982a) Late-quaternary shift of Fagus distribution. J Plant Res 95:203–217
Tsukada M (1982b) Cryptomeria japonica: glacial refugia and late-glacial and postglacial migration. Ecology 63:1091–1105
Tsukada M (1988) Japan. In: Huntley B, Webb T III (eds) Vegetation history. Kluwer, London, pp 459–518
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Yasuda Y, Miyoshi N (1998) Vegetation history in the Japanese archipelago. Asakura, Tokyo (in Japanese)
Acknowledgments
We thank Dr. Noriyuki Fujii, Dr. Naoki Nishimura, Dr. Koji Yonekura, Mr. Kozo Shibata, Dr. Tomoko Fukuda, Dr. Wataru Shinohara, Dr. Hirotoshi Sato, Dr. Hiroaki Setoguchi, Dr. Makoto Mochida, Mr. Tomoki Kadokawa and Dr. Hiroshi Ikeda for their assistance with the collection of plant materials. We also thank Dr. Takashi Sugawara, Dr. Hidetoshi Kato, Mrs. Saeko Kato, Dr. Ikuyo Saeki, Dr. Yasuyuki Watano, Dr. Vatanaparast Mohammad, and anonymous reviewers for their valuable advices. This study was partly supported by Research Project ‘A new cultural and historical exploration into human-nature relationships in the Japanese archipelago’ of the Research Institute for Humanity and Nature and Grant-in-Aid for JSPS Fellows 19-5749 to TI.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iwasaki, T., Aoki, K., Seo, A. et al. Comparative phylogeography of four component species of deciduous broad-leaved forests in Japan based on chloroplast DNA variation. J Plant Res 125, 207–221 (2012). https://doi.org/10.1007/s10265-011-0428-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-011-0428-8