Abstract
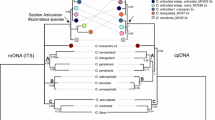
ITS sequence data were used to estimate the phylogeny of 24 Japanese Eleocharis species and to make karyomorphological observations on 19 of these taxa. Two major clades were identified in Japanese Eleocharis molecular phylogenetic trees: (1) one including all species of section Limnochloa, and (2) another comprising two sections, Pauciflorae and Eleocharis. Phylogenetic analysis including both Japanese and North American species also shows strong support for monophyly of the Mutatae/Limnochloa clade. The width of the spikelets in the species Mutatae/Limnochloa is the same as that of the culms, indicating that the relative widths of spikelets and culms are useful characteristics for classification. Two major clades were supported by karyomorphological data. All taxa of section Limnochloa had very small chromosomes, while sections Pauciflorae and Eleocharis had large chromosomes. The basic chromosome number of sections Eleocharis and Pauciflorae is thought to be x=5. Chromosomal evolution in the genus Eleocharis with diffuse centromeric chromosomes may be caused by both aneuploidization and polyploidization. Our data suggest that a 3-bp insertion near the 3′ end of the 5.8S gene is useful for intrageneric delimitations of the genus Eleocharis.






Similar content being viewed by others
References
Adachi J, Hasegawa M (1996) Computer science monographs 28. Molphy version 2.3. Programs for molecular phylogenetics base on maximum likelihood. Institute of Statistical Mathematics, Tokyo
Bruhl JJ (1995) Sedge genera of the world: relationships and a new classification of the Cyperaceae. Aust J Syst Bot 8:125–305
Clarke CB (1909) Illustrations of Cyperaceae. London
Felsenstein J (1980–2001) PHYLIP (Phylogeny Inference Package) and manual, version 3.573c. Dept Genetics, Univ of Washington, Seattle
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Goetghebeur P (1985) Studies in Cyperaceae 6. Nomenclature of the suprageneric taxa in the Cyperaceae. Taxon 34:617–632
Goldman N, Anderson JP, Rodrigo AG (2000) Likelihood-based tests of topologies in phylogenetics. Syst Biol 49:652–670
González-Elizondo MS, Peterson PM (1997) A classification of and key to the supraspecific taxa in Eleocharis. Taxon 46:433–449
Håkansson A (1954) Meiosis and pollen mitosis in x-rayed and untreated spikelets of Eleocharis palustris. Hereditas 40:325–345
Harms LJ (1968) Cytotaxonomic studies in Eleocharis subser. palustres: central united taxa. Am J Bot 55:966–974
Hasegawa M, Kishino H (1994) Accuracies of the simple methods for estimating the bootstrap probability of a maximum-likelihood tree. Mol Biol Evol 11:142–145
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Hodkinson TR, Chase MW, Lledo D, Salamin N, Renvoize SA (2002) Molecular phylogeny of Miscanthus s.l., Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) using DNA sequences from the ITS nuclear ribosomal DNA and the plastid trnL–F regions. J Plant Res 115:381–392
Hoshino T (1987) Karyomorphological studies on 6 taxa of Eleocharis in Japan. Bull Okayama Univ Sci 22A:305–312
Hoshino T, Rajbhandari KR, Ohba H (2000) Cytological studies of eleven species of Cyperaceae collected from central Nepal. Cytologia 65:219–224
Hsiao C, Chatterton NJ, Asay KH, Jensen KB (1994) Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome 37:112–120
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Kishino H, Miyata T, Hasegawa M (1990) Maximum likelihood inference of protein phylogeny and the origin of the chloroplasts. J Mol Evol 31:151–160
Koyama T (1961) Classification of the family Cyperaceae 1. J Fac Sci Univ Tokyo Sect III Bot 8:84–99
Kukkonen I (1998) Cyperaceae. In: Rechinger KH (ed) Flora Iranica, vol 173. Akademische Druck- u. Verlagsanstalt, Graz
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: Molecular Evolutionary Genetics Analysis software, Bioinformatics
Muasya AM, Simpson DS, Chase MW, Culham A (1998) An assessment of suprageneric phylogeny in Cyperaceae using rbcL DNA sequences. Plant Syst Evol 211:257–271
Nijalingappa BHM (1973) Cytological studies in Eleocharis. Caryologia 26:513–520
Ohwi J (1943) Cyperaceae Japonicae. II. A synopsis of the Rhynchosporoideae and Sciropoideae of Japan, including the Kuriles, Saghalien, Korea and Formosa. Mem Coll Sci Kyoto Imp Univ Ser B Biol 18:29–49
Olsen GJ, Matsuda H, Hagstrom R, Overbeek R (1994) FastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci 10:41–48
Rivadavia F, Kondo K, Kato M, Hasebe M (2003) Phylogeny of the sundews, Drosera (Droseraceae), based on chloroplast rbcL and nuclear 18S ribosomal DNA sequences. Am J Bot 90:123–130
Roalson EH, Friar EA (2000) Infrageneric classification of Eleocharis (Cyperaceae) revisited:evidence from the internal transcribed spacer (ITS) region of nuclear ribosomal DNA. Syst Bot 25:323–336
Roalson EH, Columbus JT, Friar EA (2001) Phylogenetic relationships in Cariceae (Cyperaceae) based on ITS (nrDNA) and trnT-L-F (cpDNA) region sequences: assessment of subgeneric and sectional relationships in Carex with emphasis on section Acrocystis. Syst Bot 26:318–341
Shimodaira H (2000) Another calculation of the p-value for the problem of regions using the scaled bootstrap resamplings. Tech Rep No 2000–35. Stanford Univ, California
Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Syst Biol 51:492–508
Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247
Simpson DA, Furness CA, Hodkinson TR, Muthama Muasya A, Chase MW (2003) Phylogenetic relationships in Cyperaceae subfamily Mapanioideae inferred from pollen and plastid DNA sequence data. Am J Bot 90:1071–1086
Smith SG, Bruhl JJ, González-Elizondo MS, Menapace FJ (2002) Eleocharis. In: Flora of North America editorial committee (ed) Flora of North America, vol 23. Magnoliophyta: Commeliaidae (in part) Cyperaceae. Oxford University Press, New York, pp 60–120
Starr JR, Bayer RJ, Ford BA (1999) The phylogenetic position of Carex section Phyllostachys and ITS implication for phylogeny and subgeneric circumscription in Carex (Cyperaceae). Am J Bot 86:563–577
Strimmer K, von Haeseler A (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol 13:964–969
Svenson HK (1929) Monographic studies in the genus Eleocharis. Rhodora 31:121–135; 167–191
Svenson HK (1934) Monographic studies in the genus Eleocharis. Rhodora 36:377–389
Svenson HK (1937) Monographic studies in the genus Eleocharis. Rhodora 39:271–272
Svenson HK (1939) Monographic studies in the genus Eleocharis. Rhodora 41:3–19; 95–104
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b 10. Sinauer Associates, Sunderland, Massachusetts
Tanaka N (1948) The problem of aneuploidy. Biological contribution in Japan (in Japanese), 4. Hokuryukan, Tokyo, pp 136–317
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tsubota H, Akiyama H, Yamaguchi T, Deguchi H (2001) Molecular phylogeny of the genus Trismegistia and related genera (Sematophyllaceae, Musci) based on chloroplast rbcL sequences. Hikobia 13:529–549
Tsubota H, Ageno Y, Estébanez B, Yamaguchi T, Deguchi H (2003) Molecular phylogeny of the Grimmiales (Musci) based on chloroplast rbcL sequences. Hikobia 14:55–70
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, New York, pp 315–22
Acknowledgements
The authors thank Dr. Marcia J. Waterway for critical review and invaluable comments on the paper. We also thank Dr. Eisuke Hayasaka, Dr. Hiroshi Ikeda, Kisaku Kameyama, Masaaki Komizunai, Dr. Masatsugu Yokota, Satoko Ozaki, Susumu Mitani, and Tomomi Masaki for their great help on field trips and in collecting plant materials.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yano, O., Katsuyama, T., Tsubota, H. et al. Molecular phylogeny of Japanese Eleocharis (Cyperaceae) based on ITS sequence data, and chromosomal evolution. J Plant Res 117, 409–419 (2004). https://doi.org/10.1007/s10265-004-0173-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-004-0173-3



