Abstract
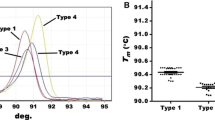
Current hepatitis C virus (HCV) genotyping techniques are often highly technical, costly, or need improvements in sensitivity and specificity. These limitations indicate the need of novel methods for HCV genotyping. The present study aimed to develop a novel genotyping method combining high-resolution melting (HRM) analysis with Bayes discriminant analysis (BDA). Target gene fragment including 5′-untranslated and core region was selected. Four or five inner amplicons for every serum were amplified using nested PCR, HRM was used to determine the melting temperature of the amplicons, and HCV genotypes were then analyzed utilizing BDA. In initial genotyping (HCV genotypes were classified into 1b, 2a, 3a, 3b, and 6a), both the overall accuracy rate and the cross-validation accuracy rate were 92.6 %, external validation accuracy rate was 95.0 %. To enhance the accuracy rate of genotyping, HCV genotypes were firstly classified into 1b, 3a, 3b, and 2a–6a, followed by a supplementary genotyping for 2a–6a. Both the overall accuracy rate and the cross-validation accuracy rate reached 97.5 %, and external validation accuracy rate was 100 %. Comparing adjusted HRM genotyping with type-specific probe technique, the difference in accuracy rates was not significant. However, the limit of detection and cost were lower for HRM. Comparing with sequencing, the limit detection of HRM was the same as the former, but the cost of HRM was lower. Hence, HRM combined with BDA was a novel method that equipped with superior accuracy, high sensitivity, and lower cost and therefore could be a better technique for HCV genotyping.





Similar content being viewed by others
References
Mukherjee R, Burns A, Rodden D, et al. Diagnosis and Management of Hepatitis C Virus Infection. J Lab Autom. 2015;20:519–38.
Lee MH, Yang HI, Yuan Y, L’Italien G, Chen CJ. Epidemiology and natural history of hepatitis C virus infection. World J Gastroenterol. 2014;20:9270–80.
Smith DB, Bukh J, Kuiken C, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–27.
Cho EJ, Jeong SH, Han BH, et al. Hepatitis C virus (HCV) genotypes and the influence of HCV subtype 1b on the progression of chronic hepatitis C in Korea: a single center experience. Clin Mol Hepatol. 2012;18:219–24.
Antaki N, Craxi A, Kamal S, et al. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int. 2010;30:342–55.
Nabi SG, Zaffar G, Sheikh NI, Hassan K, Hassan U. Hepatitis C virus genotypes: a plausible association with viral loads. Indian J Pathol Microbiol. 2013;56:384–7.
Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142–54.
Ampuero J, Romero-Gomez M, Reddy KR. Review article: HCV genotype 3—the new treatment challenge. Aliment Pharmacol Ther. 2014;39:686–98.
European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C 2014. J Hepatol. 2014;61:373–95.
World Health Organization. Guidelines for the screening, care and treatment of persons with hepatitis C infection. Geneva: World Health Organization; 2014.
Richter SS. Laboratory assays for diagnosis and management of hepatitis C virus infection. J Clin Microbiol. 2002;40:4407–12.
Halfon P, Trimoulet P, Bourliere M, et al. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J Clin Microbiol. 2001;39:1771–3.
Margraf RL, Erali M, Liew M, Wittwer CT. Genotyping hepatitis C virus by heteroduplex mobility analysis using temperature gradient capillary electrophoresis. J Clin Microbiol. 2004;42:4545–51.
Nazemi A, Tazehabadi ES, Jafarpoor M, Sharifi S. A Novel and Simple Method for HCV Genotyping. Int J Mol Clin Microbiol. 2011;1:40-45.
Zhou W, Liu Y, Yuan Q, Li X. Epileptic seizure detection using lacunarity and Bayesian linear discriminant analysis in intracranial EEG. IEEE Trans Biomed Eng. 2013;60:3375–81.
Li A, Xue Y, Jin C, Wang M, Yao X. Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian discriminant method. Biochem Biophys Res Commun. 2006;350:818–24.
Huang D, Quan Y, He M, Zhou B. Comparison of linear discriminant analysis methods for the classification of cancer based on gene expression data. J Exp Clin Cancer Res. 2009;28:149.
Poulson MD, Wittwer CT. Closed-tube genotyping of apolipoprotein E by isolated-probe PCR with multiple unlabeled probes and high-resolution DNA melting analysis. Biotechniques. 2007;43:87–91.
Liu SM, Xu FX, Shen F, Xie Y. Rapid genotyping of APOA5 -1131T > C polymorphism using high resolution melting analysis with unlabeled probes. Gene. 2012;498:276–9.
Farrar JS, Palais RA, Wittwer CT. Snapback primer genotyping of the Gilbert syndrome UGT1A1 (TA)n promoter polymorphism by high-resolution melting. Clin Chem. 2011;57:1303–10.
Minucci A, Canu G, Gentile L, et al. Small amplicons high resolution melting analysis (SA-HRMA) allows successful genotyping of acid phosphatase 1 (ACP1) polymorphisms in the Italian population; 2013;416:86–91.
Tabone T, Mather DE, Hayden MJ. Temperature switch PCR (TSP): Robust assay design for reliable amplification and genotyping of SNPs. BMC Genom. 2009;10:580.
Li Y, Wang Y, Wu G, et al. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. 2012;33:415–27.
Ulloa-Montoya F, Louahed J, Dizier B, et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31:2388–95.
Vehtari A, Gelman A, Gabry J. Efficient implementation of leave-one-out cross-validation and WAIC for evaluating fitted Bayesian models. arXiv preprint arXiv:1507.04544 2015.
Cunningham EB, Applegate TL, Lloyd AR, Dore GJ, Grebely J. Mixed HCV infection and reinfection in people who inject drugs-impact on therapy. Nat Rev Gastroenterol Hepatol. 2015;12:218–30.
Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–73.
Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61:S45–57.
Acknowledgments
This work was supported by the 863 Programme of China (2012AA022605).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Wu, D., Fu, X., Wen, Y. et al. High-resolution melting combines with Bayes discriminant analysis: a novel hepatitis C virus genotyping method. Clin Exp Med 17, 325–332 (2017). https://doi.org/10.1007/s10238-016-0424-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-016-0424-3