Abstract
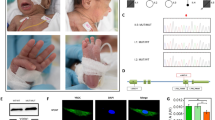
DNA repair mechanisms such as nucleotide excision repair (NER) and translesion synthesis (TLS) are dependent on proliferating cell nuclear antigen (PCNA), a DNA polymerase accessory protein. Recently, homozygosity for p.Ser228Ile mutation in the PCNA gene was reported in patients with neurodegeneration and impaired NER. Using exome sequencing, we identified a homozygous deleterious mutation, c.648delAG, in the PARP10 gene, in a patient suffering from severe developmental delay. In agreement, PARP10 protein was absent from the patient cells. We have previously shown that PARP10 is recruited by PCNA to DNA damage sites and is required for DNA damage resistance. The patient cells were significantly more sensitive to hydroxyurea and UV-induced DNA damage than control cells, resulting in increased apoptosis, indicating DNA repair impairment in the patient cells. PARP10 deficiency joins the long list of DNA repair defects associated with neurodegenerative disorders, including ataxia telangiectasia, xeroderma pigmentosum, Cockayne syndrome, and the recently reported PCNA mutation.



Similar content being viewed by others
References
Rass U, Ahel I, West SC (2007) Defective DNA repair and neurodegenerative disease. Cell 130:991–1004
McKinnon PJ (2009) DNA repair deficiency and neurological disease. Nat Rev Neurosci 10:100–112
O’Driscoll M (2012) Diseases associated with defective responses to DNA damage. Cold Spring Harb Perspect Biol 4(12):a012773
Hedglin M, Benkovic SJ (2015) Regulation of Rad6/Rad18 activity during DNA damage tolerance. Annu Rev Biophys 44:207–228
Chang DJ, Cimprich KA (2009) DNA damage tolerance: when it’s OK to make mistakes. Nat Chem Biol 5:82–90
Shivji KK, Kenny MK, Wood RD (1992) Proliferating cell nuclear antigen is required for DNA excision repair. Cell 69:367–374
Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129:665–679
Goodman MF, Woodgate R (2013) Translesion DNA polymerases. Cold Spring Harb Perspect Biol 5(10):a010363
Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141
Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14:491–500
Baple EL, Chambers H, Cross HE, et al. (2014) Hypomorphic PCNA mutation underlies a human DNA repair disorder. J Clin Invest 124:3137–3146
Nicolae CM, Aho ER, Vlahos AH, Choe KN, De S, Karras GI, Moldovan GL (2014) The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J Biol Chem 289:13627–13637
Papamichos-Chronakis M, Peterson CL (2013) Chromatin and the genome integrity network. Nat Rev Genet 14:62–67
Feijs KL, Verheugd P, Lüscher B (2013) Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J 280:3519–3529
Kleine H, Poreba E, Lesniewicz K, et al. (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32:57–69
Green CM, Baple EL, Crosby AH (2014) PCNA mutation affects DNA repair not replication. Cell Cycle 13:3157–3158
Acknowledgments
We would like to thank Drs. Kristin Eckert, Thomas Spratt, and Sergei Grigoryev for materials, support, and advice. This work was supported by the following: NIH 1R01ES026184, Department of Defense CA140303, St. Baldrick Foundation, Concern Foundation, Gittlen Foundation to GLM, and American Cancer Society ACS-IRG-13-043-01 to CMN.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The study was performed with the approval of the ethical committees of Hadassah Medical Center and the Israeli Ministry of Health.
Additional information
Maher Awni Shahrour and Claudia M. Nicolae contributed equally to this work.
An erratum to this article is available at http://dx.doi.org/10.1007/s10048-017-0511-y.
An erratum to this article is available at http://dx.doi.org/10.1007/s10048-016-0503-3.
Electronic supplementary material
ESM 1
(DOCX 19 kb)
Rights and permissions
About this article
Cite this article
Shahrour, M.A., Nicolae, C.M., Edvardson, S. et al. PARP10 deficiency manifests by severe developmental delay and DNA repair defect. Neurogenetics 17, 227–232 (2016). https://doi.org/10.1007/s10048-016-0493-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-016-0493-1