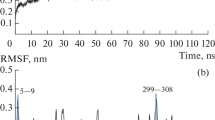
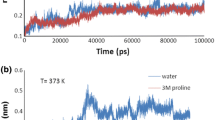
Abstract
The arginine-binding protein (ArgBP) from the hyperthermophilic eubacterium Thermotoga maritima (TmArgBP) is responsible for arginine transport through the bacterial cell membrane. The protein binds a single molecule of l-arginine, which results in conformational changes due to hinge bending. Thereby, TmArgBP acquires one of two possible conformations: open (without the presence of the arginine ligand) and closed (in the presence of the arginine ligand). Here we report a molecular dynamics study of the influence of the presence or absence of the ligand on the dynamics of TmArgBP, using the coarse-grained UNRES force field. The results of our studies indicate that binding of the arginine ligand promotes a closed conformation, which agrees with experimental data. However, the sensitivity of the TmArgBP conformation to the presence of arginine decreases and the protein becomes more flexible with increasing temperature, which might be related to the functionality of this protein in the thermophilic organism T. maritima.









Similar content being viewed by others
References
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–64. doi:10.1128/MMBR. 00031-07
Holland IB, Blight MA (1999) ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol 293:381–399. doi:10.1006/jmbi.1999.2993
Saurin W, Hofnung M, Dassa E (1999) Getting in or out: early segregation between importers and exporters in the evolution of ATP- binding cassette (ABC) transporters. J Mol Evol 48:22–41. doi:10.1007/PL00006442
Tam R, Saier MH (1993) Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev 57:320–46
Brune M, Hunter JL, Corrie JET, Webb MR (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment ATPase. Biochemistry 33:8262–8271. doi:10.1021/bi00193a013
Marvin JS, Corcoran EE, Hattangadi NA et al (1997) The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc Natl Acad Sci USA 94:4366–4371. doi:10.1073/pnas.94.9.4366
Tolosa L, Gryczynski I, Eichhorn LR et al (1999) Glucose sensor for low-cost lifetime-based sensing using a genetically engineered protein. Anal Biochem 267:114–20. doi:10.1006/abio.1998.2974
Dattelbaum JD, Lakowicz JR (2001) Optical determination of glutamine using a genetically engineered protein. Anal Biochem 291:89–95. doi:10.1006/abio.2001.4998
Salins LL, Goldsmith ES, Ensor MC, Daunert S (2001) A fluorescence-based sensing system for the environmental monitoring of nickel using the nickel binding protein from Escherichia coli. Anal Bioanal Chem 372:174–180. doi:10.1007/s00216-001-1169-7
De Lorimier RM, Smith JJ, Dwyer MA et al (2002) Construction of a fluorescent biosensor family. Protein Sci 11:2655–2675. doi:10.1110/ps.021860.nisms
Scirè A, Marabotti A, Staiano M et al (2010) Amino acid transport in thermophiles: characterization of an arginine-binding protein in Thermotoga maritima. 2. Molecular organization and structural stability. Mol Biosyst 6:687–98. doi:10.1039/b922092e
Ruggiero A, Dattelbaum JD, Staiano M et al (2014) A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima. PLoS One 9:e96560. doi:10.1371/journal.pone.0096560
Luchansky MS, Der BS, D’Auria S et al (2010) Amino acid transport in thermophiles: characterization of an arginine-binding protein in Thermotoga maritima. Mol Biosyst 6:142–51. doi:10.1039/b908412f
Ausili A, Pennacchio A, Staiano M et al (2013) Amino acid transport in thermophiles: characterization of an arginine-binding protein from Thermotoga maritima. 3. Conformational dynamics and stability. J Photochem Photobiol B 118:66–73. doi:10.1016/j.jphotobiol.2012.11.004
Deacon LJ, Billones H, Galyean AA et al (2014) Tryptophan-scanning mutagenesis of the ligand binding pocket in Thermotoga maritima arginine-binding protein. Biochimie 99:208–14. doi:10.1016/j.biochi.2013.12.011
Schlick T (2002) Molecular modeling and simulation: an interdisciplinary guide. Springer, New York
He Y, Liwo A, Weinstein H, Scheraga HA (2011) PDZ binding to the BAR domain of PICK1 is elucidated by coarse-grained molecular dynamics. J Mol Biol 405:298–314. doi:10.1016/j.jmb.2010.10.051
Gołaś E, Maisuradze GG, Senet P et al (2012) Simulation of the opening and closing of Hsp70 chaperones by coarse-grained molecular dynamics. J Chem Theory Comput 8:1750–1764. doi:10.1021/ct200680g
Rojas AV, Liwo A, Scheraga HA (2007) Molecular dynamics with the united-residue force field: ab initio folding simulations of multichain proteins. J Phys Chem B 111:293–309. doi:10.1021/jp065810x
He Y, Mozolewska MA, Krupa P et al (2013) Lessons from application of the UNRES force field to predictions of structures of CASP10 targets. Proc Natl Acad Sci USA 110:14936–41. doi:10.1073/pnas.1313316110
Kityk R, Kopp J, Sinning I, Mayer MP (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell 48:863–874. doi:10.1016/j.molcel.2012.09.023
Liwo A, Czaplewski C, Ołdziej S, Rojas AV, Kaźmierkiewicz R, Makowski M, Murarka RK, Scheraga HA (2008) Simulation of protein structure and dynamics with the coarse-grained UNRES force field. In: Voth G (ed) Coarse-graining of condensed phase and biomolecular systems, Chap. 8. CRC/Taylor & Francis, Boca Raton, pp 107–122
Liwo A, Czaplewski C, Pillardy J, Scheraga HA (2001) Cumulant-based expressions for the multibody terms for the correlation between local and electrostatic interactions in the united-residue force field. J Chem Phys 115:2323–2347. doi:10.1063/1.1383989
Kozłowska U, Maisuradze GG, Liwo A, Scheraga HA (2010) Physics-based side-chain-rotamer and side-chain and backbone virtual-bond-stretching potentials for coarse-grained UNRES force field. 2. Comparison with statistical potentials and implementation. J Comput Chem 31:1154–1167. doi:10.1002/jcc.21402
Sieradzan AK, Hansmann UHE, Scheraga HA, Liwo A (2012) Extension of UNRES force field to treat polypeptide chains with d-amino-acid residues. J Chem Theory Comput 8:4746–4757. doi:10.1021/ct3005563
Sieradzan AK, Niadzvedtski A, Scheraga HA, Liwo A (2014) Revised backbone-virtual-bond-angle potentials to treat the l- and d-amino acid residues in the coarse-grained united residue (UNRES) force field. J Chem Theory Comput 10:2194–2203. doi:10.1021/ct500119r
Khalili M, Liwo A, Rakowski F et al (2005) Molecular dynamics with the united-residue model of polypeptide chains. I. Lagrange equations of motion and tests of numerical stability in the microcanonical mode. J Phys Chem B 109:13785–97. doi:10.1021/jp058008o
Johansson MU, de Chateau M, Wikstrom M et al (1997) Solution structure of the albumin-binding GA module: a versatile bacterial protein domain. J Mol Biol 266:859–865. doi:10.1006/jmbi.1996.0856
Liwo A, Khalili M, Czaplewski C et al (2007) Modification and optimization of the united-residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins. J Phys Chem B 111:260–85. doi:10.1021/jp065380a
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi:10.1002/jcc.21256
Morris GM, Goodsell DS, Halliday RS et al (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Sieradzan AK (2015) Introduction of periodic boundary conditions into UNRES force field. J Comput Chem. doi:10.1002/jcc.23864
Huber R, Langworthy TA, Knig H et al (1986) Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch Microbiol 144:324–333. doi:10.1007/BF00409880
Liwo A, Khalili M, Scheraga HA (2005) Ab initio simulations of protein-folding pathways by molecular dynamics with the united-residue model of polypeptide chains. Proc Natl Acad Sci USA 102:2362–7. doi:10.1073/pnas.0408885102
Cuneo MJ, Changela A, Miklos AE et al (2008) Structural analysis of a periplasmic binding protein in the tripartite ATP-independent transporter family reveals a tetrameric assembly that may have a role in ligand transport. J Biol Chem 283:32812–32820. doi:10.1074/jbc.M803595200
Gonin S, Arnoux P, Pierru B et al (2007) Crystal structures of an extracytoplasmic solute receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding. BMC Struct Biol 7:11. doi:10.1186/1472-6807-7-11
Acknowledgments
A.G.L. is grateful for the support from the University of Gdansk within the Young Scientist Grant program (grant BMN 538-8370-B351-14). This work was also supported by the National Science Center of Poland (NCN), grant DEC-2012/06/A/ST4/00376. A.K.S. was supported by FNP START (100.2014) and Iuventus Plus (0558/IP3/2013/72). Calculations were carried out using our 488-processor Beowulf cluster at the Laboratory of Molecular Modeling, Faculty of Chemistry, University of Gdansk; the supercomputer resources at the Informatics Center of the Academic Computer Centre in Gdansk (CI TASK); and the Interdisciplinary Center of Mathematical and Computer Modeling (ICM) at the University of Warsaw.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper belongs to Topical Collection 6th conference on Modeling & Design of Molecular Materials in Kudowa Zdrój (MDMM 2014)
Rights and permissions
About this article
Cite this article
Lipska, A.G., Sieradzan, A.K., Krupa, P. et al. Studies of conformational changes of an arginine-binding protein from Thermotoga maritima in the presence and absence of ligand via molecular dynamics simulations with the coarse-grained UNRES force field. J Mol Model 21, 64 (2015). https://doi.org/10.1007/s00894-015-2609-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2609-1