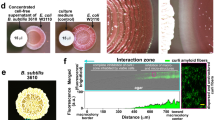
Abstract
Bacillus subtilis is one of the well-known biofilm-forming organisms associated with plants, animals, and also used as a model organism for all Bacillus sp. In B. subtilis, SrtA enzyme plays the imperative roles in mechanism of signaling pathway and microbial adherence toward the host. SrtA is highly considered as a universal drug target for all Gram positive pathogens. Because of unresolved 3D structure of SrtA in Gram positive bacteria including B. subtilis, we developed a homology model protein using structural alignments of similar SrtA from B. anthracis. While the structural model of SrtA is analyzed because of its significance in biofilm formation by screening the suitable active site based compounds and analyzing the ability of bacterial biofilm inhibition. Druggability site based screening able to retrieve the active compounds against SrtA and checked the activity of the screened compounds through experimental biochemical assays and in situ microscopic analysis. Here in this study we concluded the computationally screened SrtA inhibitors showed high level of biofilm inhibition despite difficulties in bacterial membrane rigidification. Hence this study leads a way to the new compounds that may be useful to treat the bacterial infections
Figure
Bacillus subtilis is one of the well-known biofilm-forming organisms referred to as model organism for all gram positive pathogens. From this organism, the universal drug target SrtA is evaluated for its biofilm forming ability and inhibitors are screened against the same, through in silico and in vitro methods. Druggability region based SrtA inhibitors are successful by showing anti microbial and anti biofilm activity. This study shows prominent common SrtA inhibitors against the gram positive pathogens









Similar content being viewed by others
Abbreviations
- B. subtilis:
-
Bacillus subtilis
- B. anthracis:
-
Bacillus anthracis
- SrtA:
-
Sortase A
- MD simulation:
-
Molecular dynamics simulation
- CLSM:
-
Confocal laser scanning microscopy
References
Gonzalez-Pastor JE, Hobbs EC, Losick R (2003) Cannibalism by sporulating bacteria. Science 301:510–513
Hamoen LW, Venema G, Kuipers OP (2003) Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9–17
Dahl MK, Msadek T, Kunst F, Rapoport G (1992) The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem 267:14509–14514
Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R (2001) Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626
Hamon MA, Lazazzera BA (2001) The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Wipat A, Harwood CR (1999) The Bacillus subtilis genome sequence: the molecular blueprint of a soil bacterium. FEMS Microbiol Ecol 28:1–9. doi:10.1111/j.1574-6941.1999.tb00555.x
Durrett R, Miras M, Mirouze N, Narechania A, Mandic-Mulec I, Dubnau D. (2013) Genome sequence of the Bacillus subtilis biofilm-forming transformable strain PS216. Genome Announc. 1
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209
Lopez D, Vlamakis H, Kolter R (2010) Biofilms. Cold Spring Harb Perspect Biol 2:a000398
Kolter R, Greenberg EP (2006) Microbial sciences: the superficial life of microbes. Nature 441:300–302
Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J et al (2009) Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 191:3237–3247
Bryers JD (2008) Medical biofilms. Biotech bioeng 100:1–18
O’toole JE, Tuttle J R, Tuttle M E, Lowrey T, Devereaux KM, Pax G E, Rotzoll RR. (2000) U.S. Patent No. 6,130,602. Washington, DC: U.S. Patent and Trademark Office.
Mazmanian SK, Liu G, Ton-That H, Schneewind O (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763
Marraffini LA, Dedent AC, Schneewind O (2006) Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev 70:192–221
Nagórska K, Bikowski M, Obuchowski M (2007) Multicellular behaviour and production of a wide variety of toxic substances support usage of Bacillus subtilis as a powerful biocontrol agent. Acta Biochem Polo- Eng Ed 54:495
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mole Biol Rev. 73: 310-347.
Romero D, Vlamakis H, Losick R, Kolter R (2011) An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mole microbiol 80:1155–1168
Lee VT, Schneewind O (2001) Protein secretion and the pathogenesis of bacterial infections. Genes Dev 15:1725–1752
Anderson TD, Robson SA, Jiang XW, Malmirchegini GR, Fierobe HP, Lazazzera BA et al (2011) Assembly of minicellulosomes on the surface of Bacillus subtilis. Appl Environ Microbiol 77:4849–4858
Maresso AW, Schneewind O (2008) Sortase as a target of anti-infective therapy. Pharmacol Rev 60:128–141
Levesque CM, Voronejskaia E, Huang YC, Mair RW, Ellen RP, Cvitkovitch DG (2005) Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect Immun 73:3773–3777
Weiner EM, Robson S, Marohn M, Clubb RT. The Sortase A enzyme that attaches proteins to the cell wall of Bacillus anthracis contains an unusual active site architecture. J Biol Chem. 285: 23433-43.
Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ et al (2009) The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized. J Biol Chem 284:24465–24477
Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD et al (2009) Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem 284:6924–6933
Hendrickx AP, Budzik JM, Oh SY, Schneewind O (2011) Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol 9:166–176
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Mount DW. (2007) Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc. pdb top17.
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. (2007) Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. Chapter 2: Unit 2 9.
Shafreen RM, Selvaraj C, Singh SK, Pandian SK (2013) Exploration of fluoroquinolone resistance in Streptococcus pyogenes: comparative structure analysis of wild-type and mutant DNA gyrase. J Mol Recognit 26:276–285
Thangapandian S, John S, Arooj M, Lee KW (2012) Molecular dynamics simulation study and hybrid pharmacophore model development in human LTA4H inhibitor design. PLoS ONE 7:e34593
Patschull AO, Gooptu B, Ashford P, Daviter T, Nobeli I (2013) In silico assessment of potential druggable pockets on the surface of alpha1-antitrypsin conformers. PLoS ONE 7:e36612
Tripathi SK, Singh SK, Singh P, Chellaperumal P, Reddy KK, Selvaraj C (2012) Exploring the selectivity of a ligand complex with CDK2/CDK1: a molecular dynamics simulation approach. J Mol Recognit 25:504–512
Selvaraj C, Singh SK. Validation of potential inhibitors for SrtA against Bacillus anthracis by combined approach of ligand-based and molecular dynamics simulation. J Biomol Struct Dyn. DOI:10.1080/07391102.2013.818577.
Sherman W, Beard HS, Farid R (2006) Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des 67:83–84
Barreca ML, Iraci N, De Luca L, Chimirri A (2009) Induced-fit docking approach provides insight into the binding mode and mechanism of action of HIV-1 integrase inhibitors. ChemMedChem 4:1446–1456
Selvaraj C, Singh SK, Tripathi SK, Reddy KK, Rama M (2012) In silico screening of indinavir-based compounds targeting proteolytic activity in HIV PR: binding pocket fit approach. Med Chem Res 21:4060–4068
Das D, Koh Y, Tojo Y, Ghosh AK, Mitsuya H (2009) Prediction of potency of protease inhibitors using free energy simulations with polarizable quantum mechanics-based ligand charges and a hybrid water model. J Chem Inf Model 49:2851–2862
You J, Xue X, Cao L, Lu X, Wang J, Zhang L et al (2007) Inhibition of Vibrio biofilm formation by a marine actinomycete strain A66. Appl Microbiol Biotechnol 76:1137–1144
Baldassarri L, Creti R, Recchia S, Imperi M, Facinelli B, Giovanetti E et al (2006) Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J Clin Microbiol 44:2721–2727
Hell E, Giske CG, Nelson A, Romling U, Marchini G (2010) Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis. Lett Appl Microbiol 50:211–215
Johansson EM, Crusz SA, Kolomiets E, Buts L, Kadam RU, Cacciarini M et al (2008) Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB. Chem Biol 15:1249–1257
Favre-Bonte S, Kohler T, Van Delden C (2003) Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J Antimicrob Chemother 52:598–604
Sandasi M, Leonard CM, Viljoen AM (2009) The in vitro antibiofilm activity of selected culinary herbs and medicinal plants against Listeria monocytogenes. Lett Appl Microbiol 50:30–35
Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL (2002) Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother 49:973–980
Shafreen RM, Srinivasan S, Manisankar P, Pandian SK (2012) Biofilm formation by Streptococcus pyogenes: modulation of exopolysaccharide by fluoroquinolone derivatives. J Biosci Bioeng 112:345–350
Aguilar C, Vlamakis H, Losick R, Kolter R (2007) Thinking about Bacillus subtilis as a multicellular organism. Curr opi microbial 10:638–643
Branda SS, Chu F, Kearns DB, Losick R, Kolter R (2006) A major protein component of the Bacillus subtilis biofilm matrix. Mol microbial 59:1229–1238
Kolodkin-Gal I, Cao S, Chai L, Böttcher T, Kolter R, Clardy J, Losick R (2012) A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149:684–692
Zhang J, Burrows S, Gleason C, Matthews MA, Drews MJ, LaBerge M, An YH (2006) Sterilizing Bacillus pumilus spores using supercritical carbon dioxide. J Microbiol Methods 66:479
Ahmed SF, Klena J, Husain T, Monestersky J, Naguib A, Wasfy MO (2013) Genetic characterization of antimicrobial resistance of Shigella flexneri isolates from patients in Egypt and Pakistan. Ann Clin Microbiol Antimicrob 12:9
Boyanova L, Gergova G, Nikolov R, Derejian S, Lazarova E, Katsarov N, Krastev Z (2005) Activity of Bulgarian propolis against 94 Helicobacter pylori strains in vitro by agar-well diffusion, agar dilution and disc diffusion methods. J Med Microbiol 54:481–483
Acknowledgments
The authors thankfully acknowledge the CSIR for research funding and fellowship grants (Ref. No: 37(1491)/11/EMR-II). One of the authors Chandrabose Selvaraj gratefully acknowledges CSIR for the Senior Research Fellowship (SRF). JSV gratefully acknowledges the Council of Scientific and Industrial Research (CSIR), New Delhi for the financial assistance Rendered [Ref: 9/688 (0023)/2013, EMR-I]. Poonam Singh and Sanjeev Kumar Singh acknowledge CSIR-CDRI and Alagappa University for signing the Memorandum of Understanding.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 445 kb)
Rights and permissions
About this article
Cite this article
Selvaraj, C., Sivakamavalli, J., Vaseeharan, B. et al. Examine the characterization of biofilm formation and inhibition by targeting SrtA mechanism in Bacillus subtilis: a combined experimental and theoretical study. J Mol Model 20, 2364 (2014). https://doi.org/10.1007/s00894-014-2364-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2364-8