Abstract
The effects of Cu2+ binding and the utilization of different force fields when modeling the structural characteristics of α-syn12 peptide were investigated. To this end, we performed extensive temperature replica exchange molecular dynamics (T-REMD) simulations on Cu2+-bound and unbound α-syn12 peptide using the GROMOS 43A1, OPLS-AA, and AMBER03 force fields. Each replica was run for 300 ns. The structural characteristics of α-syn12 peptide were studied based on backbone dihedral angle distributions, free-energy surfaces obtained with different reaction coordinates, favored conformations, the formation of different Turn structures, and the solvent exposure of the hydrophobic residues. The findings show that AMBER03 prefers to sample helical structures for the unbound α-syn12 peptide and does not sample any β-hairpin structure for the Cu2+-bound α-syn12 peptide. In contrast, the central structure of the major conformational clusters for the Cu2+-bound and unbound α-syn12 peptide according to simulations performed using the GROMOS 43A1 and OPLS-AA force fields is a β-hairpin with Turn9-6. Cu2+ can also promote the formation of the β-hairpin and increase the solvent exposure of hydrophobic residues, which promotes the aggregation of α-syn12 peptide. This study can help us to understand the mechanisms through which Cu2+ participates in the fibrillation of α-syn12 peptide at the atomic level, which in turn represents a step towards elucidating the nosogenesis of Parkinson’s disease.

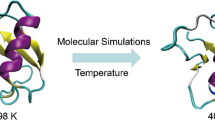
The representative structures of Cu2+-bound and unbound α-syn12 peptide using three different force fields









Similar content being viewed by others
References
Bisaglia M, Mammi S, Bubacco L (2009) FASEB J 23:329–340
Yoon J, Jang S, Lee K, Shin S (2009) J Biomol Struct Dyn 27:259–270
Yoshiki Y, Masami M, Hiroaki S, Takashi N, Shinya H, Shin-ichi H, Koichi K, Masato H (2010) J Mol Biol 395:445–456
Uversky VN, Li J, Fink AL (2001) J Biol Chem 276:44284–44296
Rasia RM, Bertoncini CW, Marsh D, Hoyer W, Cherny D, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO (2005) Proc Natl Acad Sci USA 102:4294–4299
Paik SR, Shin HJ, Lee JH, Chang CS, Kim J (1999) Biochem J 340(Pt 3):821–828
Bharathi, Rao KS (2007) Biochem Biophys Res Commun 359:115–120
Sung YH, Rospigliosi C, Eliezer D (2006) Biochim Biophys Acta 1764:5–12
Drew SC, Leong SL, Pham CL, Tew DJ, Masters CL, Miles LA, Cappai R, Barnham KJ (2008) J Am Chem Soc 130:7766–7773
Jackson MS, Lee JC (2009) Inorg Chem 48:9303–9307
Binolfi A, Rodriguez EE, Valensin D, D’Amelio N, Ippoliti E, Obal G, Duran R, Magistrato A, Pritsch O, Zweckstetter M, Valensin G, Carloni P, Quintanar L, Griesinger C, Fernandez CO (2010) Inorg Chem 49:10668–10679
Valensin D, Camponeschi F, Luczkowski M, Baratto MC, Remelli M, Valensin G, Kozlowski H (2011) Metallomics 3:292–302
Lee JC, Gray HB, Winkler JR (2008) J Am Chem Soc 130:6898–6899
Binolfi A, Lamberto GR, Duran R, Quintanar L, Bertoncini CW, Souza JM, Cervenansky C, Zweckstetter M, Griesinger C, Fernandez CO (2008) J Am Chem Soc 130:11801–11812
Ahmad A, Burns CS, Fink AL, Uversky VN (2012) J Biomol Struct Dyn 29:825–842
Dudzik CG, Walter ED, Millhauser GL (2011) Biochemistry 50:1771–1777
Riihimaki ES, Martinez JM, Kloo L (2007) J Phys Chem B 111:10529–10537
Miller Y, Ma B, Nussinov R (2010) Proc Natl Acad Sci USA 107:9490–9495
Rose F, Hodak M, Bernholc J (2011) Sci Rep 1:11
Matthes D, de Groot BL (2009) Biophys J 97:599–608
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) J Comput Chem 24:1999–2012
Best RB, Buchete NV, Hummer G (2008) Biophys J 95:L07–09
MacKerell AD Jr, Feig M, Brooks CL 3rd (2004) J Am Chem Soc 126:698–699
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) J Phys Chem B 105:6474–6487
Todorova N, Legge FS, Treutlein H, Yarovsky I (2008) J Phys Chem B 112:11137–11146
Piana S, Lindorff-Larsen K, Shaw DE (2011) Biophys J 100:L47–49
Petra K, Alfonso DS, Michal O, Robert B (2012) Biophys J 102:1897–1906
Nguyen PH, Li MS, Derreumaux P (2011) Phys Chem Chem Phys 13:9778–9788
Cao Z, Wang J (2010) J Biomol Struct Dyn 27:651–661
Cao Z, Liu L, Wang J (2011) J Biomol Struct Dyn 29:527–539
Cao Z, Liu L, Zhao L, Wang J (2011) Int J Mol Sci 12:8259–8274
Hess B (2008) J Chem Theory Comput 4:116–122
van der Spoel D, van Drunen R, Berendsen HJC (1994) GRoningen MAchine for Chemical Simulations. BIOSON Research Institute, Groningen
van Gunsteren WF, Billeter SR, Eising AA, Hunenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Vdf Hochschulverlag AG an der ETH Zürich, Zürich
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J (1981) Interaction models for water in relation to protein hydration. In: Pullman B (ed) Intermolecular forces. Reidel, Dordrecht, pp 331–342
Jorgensen WL, Chandrasekhar J, Madura JD, Impy RW, Klein ML (1983) J Chem Phys 79:926–935
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) J Chem Phys 103:8577–8593
Bussi G, Donadio D, Parrinello M (2007) J Chem Phys 126:014101
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) J Chem Phys 81:3684–3690
Sugita Y, Okamoto Y (1999) Chem Phys Lett 314:141–151
Patriksson A, van der Spoel D (2008) Phys Chem Chem Phys 10:2073–2077
Cao Z, Liu L, Wu P, Wang J (2011) Acta Biochim Biophys Sinica 43:172–180
Hu H, Elstner M, Hermans J (2003) Proteins 50:451–463
Best RB, Mittal J (2010) J Phys Chem B 114:8790–8798
Heinig M, Frishman D (2004) Nucleic Acids Res 32:W500–502
Garcia AE (1992) Phys Rev Lett 68:2696–2699
Dobson CM (2003) Nature 426:884–890
Takao Y, Yuji S, Yuko O (2004) Chem Phys Lett 386:460–467
Rueda M, Ferrer-Costa C, Meyer T, Perez A, Camps J, Hospital A, Gelpi JL, Orozco M (2007) Proc Natl Acad Sci USA 104:796–801
Acknowledgments
The authors thank Prof. H.J.C. Berendsen (University of Groningen) for providing us with the GROMACS programs.
This work was supported by grants 31000324, 61271378 and 30970561 from the National Natural Science Foundation of China and grants 2009ZRA14027 and 2009ZRA14028 from the Shandong Province Natural Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1a–b
Initial structures of a the unbound and b the Cu2+-bound α-syn12 peptide. (DOC 84 kb)
Rights and permissions
About this article
Cite this article
Cao, Z., Liu, L., Zhao, L. et al. Comparison of the structural characteristics of Cu2+-bound and unbound α-syn12 peptide obtained in simulations using different force fields. J Mol Model 19, 1237–1250 (2013). https://doi.org/10.1007/s00894-012-1664-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1664-0