Abstract
The ionotropic N-methyl-d-aspartate (NMDA) receptor is of importance in neuronal development, functioning, and degeneration in the mammalian central nervous system. The functional NMDA receptor is a heterotetramer comprising two NR1 and two NR2 or NR3 subunits. We have carried out evolutionary trace (ET) analysis of forty ionotropic glutamate receptor (IGRs) sequences to identify and characterize the residues forming the binding socket. We have also modeled the ligand binding core (S1S2) of NMDA receptor subunits using the recently available crystal structure of NR1 subunit ligand binding core which shares ~40% homology with other NMDA receptor subunits. A short molecular dynamics simulation of the glycine-bound form of wild-type and double-mutated (D481N; K483Q) NR1 subunit structure shows considerable RMSD at the hinge region of S1S2 segment, where pore forming transmembrane helices are located in the native receptor. It is suggested that the disruption of domain closure could affect ion-channel activation and thereby lead to perturbations in normal animal behavior. In conclusion, we identified the amino acids that form the ligand-binding pocket in many ionotropic glutamate receptors and studied their hydrogen bonded and nonbonded interaction patterns. Finally, the disruption in the S1S2 domain conformation (of NR1 subunit- crystal structure) has been studied with a short molecular dynamics simulation and correlated with some experimental observations.
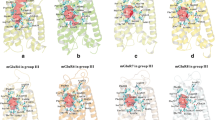
Figure The figure shows the binding mechanism of glutamate with NR2B subunit of the NMDA receptor. Glutamate is shown in cpk, hydrogen bonds in dotted lines and amino acids in blue. The amino acids shown here are within a 4-Å radius of the ligand (glutamate)










Similar content being viewed by others
References
Dingledine R, Borges K, Bowie D, Traynelis SF (1999) Pharmacol Rev 51:7–61
Cull-Candy S, Brickley S, Farrant M (2001) Curr Opin Neurobiol 11:327–335
Mayer ML, Westbrook GL, Guthrie PB (1984) Nature 309:261–263
Madison DV, Malenka RC, Nicoll RA (1991) Annu Rev Neurosci 14:379–397
Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B Seeburg PH (1992) Science 256:1217–1221
Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M (1992) Nature 358:36–41
Johnson JW, Ascher P (1987) Nature 325:529–531
Kleckner NW, Dingledine R (1988) Science 241:835–837
Laube B, Hirai H, Sturgess M, Betz H, Kuhse J (1997) Neuron 18:493–503
Kemp JA, Bluethmann H, Kew JN (2002) 22:6713–6723
Anson LC, Chen PE, Wyllie DJ, Colquhoun D, Schoepfer R (1998) J Neurosci 18:581–589
Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D (2002) Nature 415:793-798
Matsuda K, Kamiya Y, Matsuda S, Yuzaki M (2002) Brain Res Mol Brain Res 100:43–52
Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N (1998) Nature 393:377–381
Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF (2001) J Neurosci 21:1228–1237
Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA (1995) J Neurosci 15:6498–6508
Furukawa H, Gouaux E (2003) EMBO J 22:2873–2885
Armstrong N, Gouaux E (2000) Neuron 28:165–181
Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O’Hara PJ, Heinemann SF (1994) Neuron 13:1345–1357
Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, Ames GF, Kim SH (1993) J Biol Chem 268:11348–1155
Lummis SC, Fletcher EJ, Green T (2002) Neuropharmacology 42:437–443
Tikhonova IG, Baskin II, Palyulin VA, Zefirov NS (2003) J Med Chem 46:1609–1616
Tikhonova IG, Baskin II, Palyulin VA, Zefirov NS, Bachurin SO (2002) J Med Chem 45:3836–3843
Kuusinen A, Arvola M, Keinanen K (1995) EMBO J 14:6327–6332
Ivanovic A, Reilander H, Laube B, Kuhse J (1998) J Biol Chem 273:19933–19937
Keinanen K, Jouppila A, Kuusinen A (1998) Biochem J 330:1461–1467
Lichtarge O, Bourne HR, Cohen FE (1996) Proc Natl Acad Sci USA 93:7507–7511
Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T (1994) Neuron 13:325–338
Miyamoto Y, Yamada K, Noda Y, Mori H, Mishina M, Nabeshima T (2001) J Neurosci 21:750–757
Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S (1994) Cell 76:427–437
Mohn AR, Gainetdinov RR, Caron MG, Koller BH (1999) Cell 98:427–436
Kiefer F, Jahn H, Koester A, Montkowski A, Reinscheid RK, Wiedemann K (2003) Biol Psychiatry 53:345–351
Ballard TM, Pauly-Evers M, Higgins GA, Ouagazzal AM, Mutel V, Borroni E, Kemp JA, Bluethmann H, Kew JN (2002) J Neurosci 22:6713–6723
Kew JN, Koester A, Moreau JL, Jenck F, Ouagazzal AM, Mutel V, Richards JG, Trube G, Fischer G, Montkowski A, Hundt W, Reinscheid RK, Pauly-Evers M, Kemp JA, Bluethmann H (2000) J Neurosci 20:4037–4049
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25:3389–3402
Rost B, Sander C (1993) J Mol Biol 232:584–599
Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acids Res 22:4673–4680
Sali A, Blundell TL (1993) J Mol Biol 234:779–815
Luthy R, Bowie JU, Eisenberg D (1992) Nature 356:83–85
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) Jol Appl Crystal 26:283–291
All tools utilized herein were accessed and utilized as implemented in Insight II-97.5, Accelrys (http://www.accelrys.com)
Kleywegt GJ, Jones TA (1997) Methods Enzymol 277:525–545
Guex N, Peitsch MC (1997) Electrophoresis 18:2714–2723
Sayle RA, Milner-White EJ (1995) Trends Biochem Sci 20:374
Chothia C, Lesk AM (1986) EMBO J 5:823–826
Zvelebil MJ, Barton GJ, Taylor WR, Sternberg MJ (1987) J Mol Biol 195:957–961
Baldwin JM (1993) EMBO J 12:1693–1703
Zvelebil MJ, Sternberg MJ (1988) Protein Eng 2:127–138
Arinaminpathy Y, Biggin PC, Shrivastava IH, Sansom MS (2003) FEBS Lett 553:321–327
Arinaminpathy Y, Sansom MS, Biggin PC (2002) Biophys J 82:676–683
Acknowledgements
We thank Dr. Chittaranjan Andrade and Dr. Innis Axel for language correction of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Blaise, MC., Sowdhamini, R., Rao, M.R.P. et al. Evolutionary trace analysis of ionotropic glutamate receptor sequences and modeling the interactions of agonists with different NMDA receptor subunits. J Mol Model 10, 305–316 (2004). https://doi.org/10.1007/s00894-004-0196-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-004-0196-7