Abstract
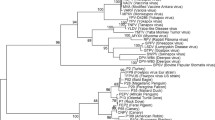
Here, we report the genome sequence of a feral pigeon alphaherpesvirus (columbid herpesvirus type 1, CoHV-1), strain HLJ, and compare it with other avian alphaherpesviruses. The CoHV-1 strain HLJ genome is 204,237 bp in length and encodes approximately 130 putative protein-coding genes. Phylogenetically, CoHV-1 complete genome resides in a monophyletic group with the falconid herpesvirus type 1 (FaHV-1) genome, distant from other alphaherpesviruses. Interestingly, the evolutionary analysis of partial genes of CoHV-1 isolated from different organisms and areas (currently accessible on GenBank) indicates that the CoHV-1 HLJ strain isolated from pigeon (Columba livia) is closely related to the strains isolated from peregrine falcon (Falco peregrinus) in Poland and owl (Bubo virginianus) in USA. These results may suggest possible transmission of the virus between different organisms and different geographic areas.


Similar content being viewed by others
References
Smadel JE, Jackson EB, Harman JW (1945) A new virus disease of pigeons: recovery of the virus. J Exp Med 81:385–398
Gailbreath KL, Oaks JL (2008) Herpesviral inclusion body disease in owls and falcons is caused by the pigeon herpesvirus (columbid herpesvirus 1). J Wildl Dis 44:427–433
Pinkerton ME, Wellehan JJ, Johnson AJ, Childress AL, Fitzgerald SD, Kinsel MJ (2008) Columbid herpesvirus-1 in two Cooper’s hawks (Accipiter cooperii) with fatal inclusion body disease. J Wildl Dis 44:622–628
Wozniakowski GJ, Samorek-Salamonowicz E, Szymanski P, Wencel P, Houszka M (2013) Phylogenetic analysis of Columbid herpesvirus-1 in rock pigeons, birds of prey and non-raptorial birds in Poland. BMC Vet Res 9:52
Zhao PP, Ma J, Guo Y, Tian L, Guo GY, Zhang KX, Xing MW (2015) Isolation and characterization of a herpesvirus from feral pigeons in China. Vet J 206:417–419
Goldenberger D, Perschil I, Ritzler M, Altwegg M (1995) A simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl 4:368–370
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Huang W, Xie P, Xu M, Li P, Zao G (2011) The influence of stress factors on the reactivation of latent herpes simplex virus type 1 in infected mice. Cell Biochem Biophys 61:115–122
Wang J, Hoper D, Beer M, Osterrieder N (2011) Complete genome sequence of virulent duck enteritis virus (DEV) strain 2085 and comparison with genome sequences of virulent and attenuated DEV strains. Virus Res 160:316–325
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or personal relationships with other people or organizations that could inappropriately influence or bias the content of this paper.
Ethical approval
All procedures used in this study were approved by the Institution Animal Care and Use Committee of Northeast Forestry University (UT-17; 25 July 2012).
Funding
This study was provided by the National Natural Science Foundation of China (Grant No. 31672619), the Fundamental Research Funds for the Central Universities (Grant No. 2572016EAJ5) and the Natural Science Foundation of Heilongjiang Province (Grant No. C2015061).
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3329_MOESM1_ESM.tif
Supplementary Figure 1: Genetic organization of the CoHV-1 genome. This map shows the class E genome structure of CoHV-1 and indicates the location, direction and sizes of annotated ORFs, numbered based on the CoHV-1 locus tag and labeled according to homology with the characterized HHV-1 genome as well as other avian herpesviruses. The genomic genes were numbered from 5’ to 3’ accordant with the direction of the black arrows in the map. (TIFF 6115 kb)
Rights and permissions
About this article
Cite this article
Guo, Y., Li, S., Sun, X. et al. Complete genome sequence and evolution analysis of a columbid herpesvirus type 1 from feral pigeon in China. Arch Virol 162, 2131–2133 (2017). https://doi.org/10.1007/s00705-017-3329-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3329-x