Abstract
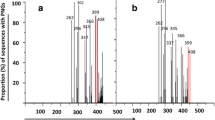
Pegylated interferon and ribavirin combination therapy effectively suppresses viral replication in 50 %–60 % of hepatitis C virus (HCV)-infected patients. However, HCV-infected patients often display varied responses to therapy, and strains of subtype lb (the most widespread HCV subtype worldwide) have more-severe clinical manifestations, greater viral loads, and poorer responses to interferon treatment. Therefore, understanding the genomic variability of HCV is crucial to treatment of HCV infection. In this study, we used the appropriate software to analyze the nucleotide, and amino acid sequences of the envelope proteins (E1 and E2) of HCV to investigate the extent of their variability in several HCV subtypes (1a, 1b, 2a, 2b, 3a, 4a, 5a and 6a) and calculated the ratio of nonsynonymous to synonymous substitutions (dN/dS) in these proteins to investigate the immunological pressure acting on them. We also predicted the N-glycosylation sites in E1 and E2 to determine their association with viral neutralization. We found that E1 is more variable, has a higher dN/dS ratio, and has more N-glycosylation sites than E2 in HCV subtype 1b. This indicates that the variability of E1, its dN/dS ratio, and its degree of N-glycosylation might play an important role in the treatment of infection with HCV subtype 1b.


Similar content being viewed by others
References
Steinhauer DA, Domingo E et al (1992) Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122(2):281–288
Bruno S, Silini E et al (1997) Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a prospective study. Hepatology 25(3):754–758
Tanaka H, Tsukuma H et al (1998) Hepatitis C virus 1b(II) infection and development of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma: a case-control study in Japan. J Epidemiol 8(4):244–249
Simmonds P (2004) Genetic diversity and evolution of hepatitis C virus—15 years on. J Gen Virol 85(Pt 11):3173–3188
Simmonds P, Bukh J et al (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42(4):962–973
Ray SC, Wang YM et al (1999) Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol 73(4):2938–2946
Xu CH, Shen T et al (2012) Higher dN/dS ratios in the HCV core gene, but not in the E1/HVR1 gene, are associated with human immunodeficiency virus-associated immunosuppression. Arch Virol 157(11):2153–2162
Thompson JD, Gibson TJ et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882
Hussein N, Zekri AN et al (2014) New insight into HCV E1/E2 region of genotype 4a. Virol J 11(1):2512
Xiao F, Fofana I et al (2015) Synergy of entry inhibitors with direct-acting antivirals uncovers novel combinations for prevention and treatment of hepatitis C. Gut 64(3):483–494
Nayak A, Pattabiraman N et al (2015) Structure–function analysis of hepatitis C virus envelope glycoproteins E1 and E2. J Biomol Struct 33:1682–1694
Sullivan DG, Wilson JJ et al (1998) Multigene tracking of hepatitis C virus quasispecies after liver transplantation: correlation of genetic diversification in the envelope region with asymptomatic or mild disease patterns. J Virol 72(12):10036–10043
Naderi M, Gholipour N et al (2014) Hepatitis C virus and vaccine development. Int J Mol Cell Med 3(4):207–215
Netski DM, Mao Q et al (2008) Genetic divergence of hepatitis C virus: the role of HIV-related immunosuppression. J Acquir Immune Defic Syndr 49(2):136–141
Kachko A, Kochneva G et al (2011) New neutralizing antibody epitopes in hepatitis C virus envelope glycoproteins are revealed by dissecting peptide recognition profiles. Vaccine 30(1):69–77
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
X.-D. Cheng and H.-F. Xu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cheng, XD., Xu, HF., Wei, XM. et al. Variation analysis of E1 and E2 in HCV subtypes. Arch Virol 160, 2479–2482 (2015). https://doi.org/10.1007/s00705-015-2533-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2533-9