Abstract
Background
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a powerful tool to assess the diversity of a microbial community. In this study, we performed T-RFLP analysis of the fecal microbiota from patients with ulcerative colitis (UC) and those with Crohn’s disease (CD).
Methods
Thirty-one patients with UC, 31 patients with CD, and 30 healthy individuals were enrolled. The polymerase chain reaction (PCR) products obtained from the 16S rRNA genes of fecal samples were digested with BslI, and T-RF lengths were determined.
Results
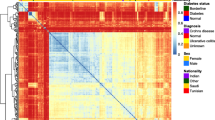
The fecal microbial communities were classified into 5 clusters. Twenty-eight of the 30 healthy individuals and 17 of the 18 patients with inactive UC were classified into clusters I, II, and III, but these clusters included a small number of patients with active UC and inactive/active CD. In contrast, 8 of the 13 patients with active UC and the majority of CD patients (12 of the 16 patients with inactive CD, and 11 of the 15 patients with active CD) were included in clusters IV and V. Based on the BslI-digested T-RFLP database, the bacteria showed a significant decrease in the Clostridium family in patients with active UC and inactive/active CD. In contrast, Bacteroides were significantly increased in CD patients. No significant differences were observed between patients with active UC and those with active CD.
Conclusion
The fecal microbial communities of IBD patients were different from those of healthy individuals. The gut microbiota of patients with inactive UC tended to be closer to that of healthy individuals, suggesting different roles for the fecal microbiota in the pathophysiology of UC and CD.

Similar content being viewed by others
References
Sands BE. Inflammatory bowel disease: past, present, and future. J Gastroenterol. 2007;42:16–25.
Mayer L. Evolving paradigms in the pathogenesis of IBD. J Gastroenterol. 2010;45:9–16.
Hibi T, Ogata H. Novel pathophysiological concepts of inflammatory bowel disease. J Gastroenterol. 2006;41:10–6.
Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29.
Mizoguchi A, Mizoguchi E. Inflammatory bowel disease, past, present and future: lessons from animal models. J Gastroenterol. 2008;43:1–17.
Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407.
Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–83.
Braun J, Wei B. Body traffic: ecology, genetics, and immunity in inflammatory bowel disease. Annu Rev Pathol. 2007;2:401–29.
Seksik P, Sokol H, Lepage P, Vasquez N, Manichanh C, Mangin I, et al. Review article: the role of bacteria in onset and perpetuation of inflammatory bowel disease. Aliment Pharmacol Ther. 2006;24(Suppl 3):11–8.
Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–76.
Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94.
Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–4.
Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–9.
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8.
Mahida YR, Rolfe VE. Host-bacterial interactions in inflammatory bowel disease. Clin Sci (Lond). 2004;107:331–41.
Hayashi H, Sakamoto M, Kitahara M, Benno Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol Immunol. 2003;47:557–70.
Sakamoto M, Hayashi H, Benno Y. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol Immunol. 2003;47:133–42.
Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J Med Microbiol. 2003;52(Pt 1):79–89.
Li F, Hullar MA, Lampe JW. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J Microbiol Methods. 2007;68:303–11.
Schutte UM, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, et al. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biotechnol. 2008;80:365–80.
Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54(Pt 11):1093–101.
Andoh A, Benno Y, Kanauchi O, Fujiyama Y. Recent advances in molecular approaches to gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15:2066–73.
Marsh TL, Saxman P, Cole J, Tiedje J. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol. 2000;66:3616–20.
Andoh A, Sakata S, Koizumi Y, Mitsuyama K, Fujiyama Y, Benno Y. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–62.
Andoh A, Tsujikawa T, Sasaki M, Mitsuyama K, Suzuki Y, Matsui T, et al. Fecal microbiota profile of Crohn’s disease determined by terminal restriction fragment length polymorphism (t-rflp) analysis. Aliment Pharmacol Ther. 2009;29:75–82.
Rachmilewitz D. Coated mesalazine (5-aminosalicylic acid) versus sulphasalazine in the treatment of active ulcerative colitis: a randomised trial. BMJ. 1989;298:82–6.
Best WR, Becktel JM, Singleton JW, Kern F Jr. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology. 1976;70:439–44.
Hayashi H, Sakamoto M, Kitahara M, Benno Y. Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol Lett. 2006;257:202–7.
Nagashima K, Hisada T, Sato M, Mochizuki J. Application of new primer–enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl Environ Microbiol. 2003;69:1251–62.
Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–5.
Matsuda H, Fujiyama Y, Andoh A, Ushijima T, Kajinami T, Bamba T. Characterization of antibody responses against rectal mucosa-associated bacterial flora in patients with ulcerative colitis. J Gastroenterol Hepatol. 2000;15:61–8.
Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55(Pt 5):617–24.
Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54.
Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–48.
Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–75.
Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–807.
Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–18.
Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–45.
Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–41.
Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermu dez-Humara n LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–6.
Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2:716–27.
Andoh A, Fujiyama Y, Hata K, Araki Y, Takaya H, Shimada M, et al. Counter-regulatory effect of sodium butyrate on tumour necrosis factor-alpha (TNF-alpha)-induced complement C3 and factor B biosynthesis in human intestinal epithelial cells. Clin Exp Immunol. 1999;118:23–9.
Kawamura T, Andoh A, Nishida A, Shioya M, Yagi Y, Nishimura T, et al. Inhibitory effects of short-chain fatty acids on matrix metalloproteinase secretion from human colonic subepithelial myofibroblasts. Dig Dis Sci. 2009;54:238–45.
Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–51.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andoh, A., Imaeda, H., Aomatsu, T. et al. Comparison of the fecal microbiota profiles between ulcerative colitis and Crohn’s disease using terminal restriction fragment length polymorphism analysis. J Gastroenterol 46, 479–486 (2011). https://doi.org/10.1007/s00535-010-0368-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-010-0368-4