Abstract
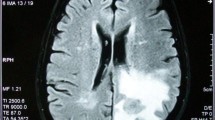
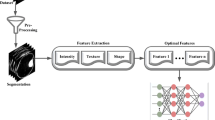
This paper presents a new classification technique for 3D MR images, based on a third-generation network of spiking neurons. Implementation of multi-dimensional co-occurrence matrices for the identification of pathological tumor tissue and normal brain tissue features are assessed. The results show the ability of spiking classifier with iterative training using genetic algorithm to automatically and simultaneously recover tissue-specific structural patterns and achieve segmentation of tumor part. The spiking network classifier has been validated and tested for various real-time and Harvard benchmark datasets, where appreciable performance in terms of mean square error, accuracy and computational time is obtained. The spiking network employed Izhikevich neurons as nodes in a multi-layered structure. The classifier has been compared with computational power of multi-layer neural networks with sigmoidal neurons. The results on misclassified tumors are analyzed and suggestions for future work are discussed.















Similar content being viewed by others
References
American Cancer Society (2012) Cancer facts & figures 2012. American Cancer Society, Atlanta
Bellatreche A, McGuire LP, McGiniity M, Wu Xiang Q (2006) Evolutionary design of spiking neural networks. New Math Nat Comput 2(3):237–253
Bohte SM, Kok JN, La Poutre H (2002) Error-backpropagation in temporally encoded networks of spiking neurons. Neurocomputing 48:17–37
Chen J, Suarez J, Molnar P, Behal A (2011) Maximum likelihood parameter estimation in a stochastic resonate-and-fire neuronal model. In: IEEE 1st international conference on computational advances in bio and medical sciences (ICCABS), pp 57–62
Chin-Chen C, Zen-Chung S, Chia-Wen C, Wen-Kai T, Der-Lor W (2013) Feature based 3D texture synthesis approach. Int J Innov Comput Inf Control 9(3):1201–1210
Deepa SN, Arunadevi B (2014) Multi-dimensional texture characterization. On analysis for brain tumor tissues using MRS and MRI. J Digit Imaging 2:377–386
Gerstner W, Kistler W (2002) Spiking neuron models: single neurons, populations, plasticity. Cambridge University Press, London
Ghodrati M, Khaligh-Razavi SM, Ebrahimpour R, Rajaei K, Pooyan M (2012) How can selection of biologically inspired features improve the performance of a robust object recognition model? PLoS One 17(2). doi:10.1371/journal.pone.003235
Ghosh-Dastidar S, Adeli H (2007) Improved spiking neural networks for EEG classification and epilepsy and seizure detection. Integr Comput Aided Eng 14(3):187–212
Goel P, Liu H, Brown D, Datta A (2008) On the use of spiking neural network for EEG classification. Int J Knowl Based Intell Eng Syst 12:295–304
Guo X, Johe K, Molnar P, Davis H, Hickman J (2010) Characterization of a human fetal spinal cord stem cell line, NSI-566RSC, and its induction to functional motoneurons. J Tissue Eng Regen Med 4(3):181–193
Gupta A, Long LN (2007) Character recognition using spiking neural networks. In: Proceedings of international joint conference on neural networks, Florida, pp 53–58
Izhikevich EM (2004) Which model to use for cortical spiking neurons? IEEE Trans Neural Netw 15:1063–1070
Jin Y, Wen R, Sendhoff B (2007) Evolutionary multi-objective optimization of spiking neural networks. In: Proceedings of ICANN. LNCS, vol 4668, pp 370–379
Kamoi S, Iwai R, Kinjo H, Ymamoto T (2003) Pulse pattern training of spiking neural networks using improved genetic algorithm. In: Proceedings of international symposium on computational intelligence in robotics and automation, Kobe, Japan, pp 977–981
Kampakis S (2012) Improved Izhikevich neurons for spiking neural networks. Soft Comput 16:943–953
Kasinski A, Ponulak F (2006) Comparison of supervised learning methods for spike time coding in spiking neural networks. Int J Appl Math Comput Sci 16(1):101–113
Kovalev VA, Petrou M (1996) Multidimensional co-occurrence matrices for object recognition and matching. Graph Models Image Process 58(3):187–197
Kovalev VA, Kruggel F, Gertz HJ, von Cramon DY (2001) Three dimensional texture analysis of MRI brain datasets. IEEE Trans Med Imaging 20:424–433
Long LN, Fang G (2010) A review of biologically plausible neuron models for spiking neural networks. In: Proceedings of AIAA InfoTech@ aerospace conference, Atlanta, GA, pp 2010–3540
Maass W (2002) Computation with spiking neurons. In: Arbib MA (ed) The handbook of brain theory and neural networks, 2nd edn. MIT Press, Cambridge, USA, pp 1080–1083
Maass W (1997b) Fast sigmoidal networks via spiking neurons. Neural Comput 9:279–279
Maass W (1997a) Networks of spiking neurons: the third generation of neural network models. Neural Netw Comput Vis Pattern Recognit 10:1659–1671
Maass W, Bishop CM (1999) Pulsed neural networks. MIT Press, Cambridge
Meftah B, Lezoray O, Benyettou A (2010) Segmentation and edge detection based on spiking neural network model. Neural Process Lett. doi:10.1007/s11063-010-9149-6
Michael R, Kaus Simon K, Warfield Nabavi A, Peter M, Ferenc A, Kikinis R (2001) Automated segmentation of MR images of brain tumors. Radiology 218:586–591
Mizoguchi N, Nagamatsu Y, Aihara K, Kohno T (2011) A two-variable silicon neuron circuit based on the Izhikevich model. Artif Life Robot 16:383–388
O’Halloran M, McGinley B, Conceicao RC, Morgan F, Jones E, Glavin M (2011) Spiking neural networks for breast cancer classification in a dielectrically heterogeneous breast. Prog Electromagn Res 113:413–428
Panchev C, Wermter S (2006) Temporal sequence detection with spiking neurons: towards recognizing robot language instruction. Connect Sci 18:1–22
Russell A, Orchard G, Dong Y, Mihalas S, Niebur E, Tapson J, Etienne-Cummings R (2010) Optimization methods for spiking neurons and networks. IEEE Trans Neural Netw 2(12):1950–1962
Sivanandam SN, Deepa SN (2005) Introduction to soft computing, 2nd edn. Wiley Publishers, New Delhi
Stromatias E (2011) Developing a supervised training algorithm for limited precision feed-forward spiking neural networks. MS Thesis, University of Liverpool
Vazquez RA (2010a) Pattern recognition using spiking neurons and firing rates. In: Kuri-Morales A, Simari GR (eds) IBERAMIA 2010. LNCS, vol 6433, pp 423–432
Vazquez RA, Garro BA (2011)Training spiking neurons by means of particle swarm optimization. In: Proceedings of ICSI 2011, Part I. LNCS, vol 6728, pp 242–249
Vazquez RA (2010b) Izhikevich neuron model and its application in pattern recognition. Aust J Intell Inf Process Syst 11:35–40
Wu QX, McGinnity M, Maguire LP, Belatreche A, Glackin B (2008) Processing visual stimuli using hierarchical spiking neural networks. Neurocomputing 71(10):2055–2068
Wysoski SG, Benuskova L, Kasabov N (2007) Text-independent speaker authentication with spiking neural networks. In: Proceedings of international conference on artificial neural networks, Porto, Portugal, pp 758–767
Zhang J, Yu C, Jiang G, Liu W, Tong L (2012) 3D texture analysis on MRI images of Alzheimer’s disease. Brain Imaging Behav 6:61–69
Zhi L, Chen J, Molnar P, Behal A (2012) Weighted least-squares approach for identification of a reduced-order adaptive neuronal model. IEEE Trans Neural Netw Learn Syst 23(5):834–840
Zucker SW, Hummel RA (1981) A 3D edge operator. IEEE Trans Pattern Analysis 3:324–331
Acknowledgments
This work was supported by the Council of Scientific and Industrial Research (CSIR), India with reference 09/1073/ (0001)/2012. The authors would thank PSG IMSR & Hospitals, for providing clinical data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by V. Loia.
Rights and permissions
About this article
Cite this article
Baladhandapani, A., Nachimuthu, D.S. Evolutionary learning of spiking neural networks towards quantification of 3D MRI brain tumor tissues. Soft Comput 19, 1803–1816 (2015). https://doi.org/10.1007/s00500-014-1364-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-014-1364-z