Abstract
Key message
Genetic diversity of Monoon tirunelveliense is reported here from India to facilitate its conservation and to take up such studies for conserving threatened plants from different parts of the world.
Abstract
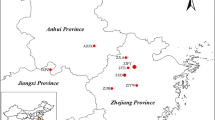
As endemic and threatened species are susceptible to loss of genetic variation due to stochastic factors, investigation was carried out to assess genetic diversity of Monoon tirunelveliense (Annonaceae), which is a steno endemic and a critically endangered species from the Kalakkad-Mundanthurai Tiger Reserve in India, using inter-simple sequence repeats (ISSRs). Thirty-five samples collected from seven populations were screened by 34 primers, wherein 11 primers produced 65 clear and reproducible bands that included 43.08 % of polymorphic bands (PPB), at the rate of 1–6 bands population−1 and 5.90 bands primer−1. Nei’s genetic diversity (H) was 0.1428, on an average. Analysis of Shannon information index (I) showed 0.2120, on an average, and total genetic diversity (H T) was moderate with 0.2651 and genetic diversity within populations (H S) was a low of 0.1428. Proportion of total diversity among populations (G ST) was 0.4613. Gene flow among populations (Nm) was 0.5839 individuals generation−1 indicating a low migration rate among populations. Mantel test showed no significant correlation among genetic differentiation and geographical distance (r = 0.489, p = 0.007). Phylogenetic relationship among Monoon species using rbcL and matK gene sequences derived by combined Bayesian analysis is given. Besides conserving all the existing populations, matured fruits should be collected and sown in debris-free soil to increase the percentage of seed germination as seeds are highly vulnerable to the attack of termites and microbial decay. Seeds from genetic diversity-rich Populations 2 and 3 are recommended for propagation to increase the extent and occurrence of the species.




Similar content being viewed by others
References
Alvarez-Buylla ER, Garcia-Barrios R, Lara-Moreno C, Martinez-Ramos M (1996) Demographic and genetic models in conservation biology: applications and perspectives for Tropical Rain Forest tree species. Annu Rev Ecol Evol Syst 27:387–421
Ashton PS (1969) Speciation among tropical forest trees: some deductions in the light of recent evidence. Biol J Linn Soc 1:155–196
Austerlitz F, Mariette S, Machon N, Gouyon P-H, Godelle B (2000) Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics 154:1309–1321
Cao PJ, Yao QF, Ding BY, Zeng HY, Zhong YX, Fu CX, Jin XF (2006) Genetic diversity of Sinojackia dolichocarpa (Styracaceae), a species endangered and endemic to China, detected by inter-simple sequence repeat (ISSR). Biochem Sys Ecol 34:231–239
Chaowasku T, Johnson DM, Van der Ham RWJM, Chatrou LW (2012) Characterization of Hubera (Annonaceae), a new genus segregated from Polyalthia and allied to Miliusa. Phytotaxa 69:33–56
Couvreur TLP, Pirie MD, Chatrou LW, Saunders RMK, Su YCF, Richardson JE, Erkens RHJ (2011) Early evolutionary history of the flowering plant family Annonaceae: steady diversification and boreotropical geodispersal. J Biogeogr 38:664–680
Culley TM, Wolfe AD (2001) Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86:545–556
Deshpande AU, Apte GS, Bahulikar RA, Lagu MD, Kulkarni BG, Suresh HS, Singh NP, Rao MKV, Gupta VS, Pant A, Ranjekar PK (2001) Genetic diversity across natural populations of three montane plant species from the Western Ghats, India revealed by inter simple sequence repeats. Mol Ecol 10:2397–2408
Doyle J (1991) DNA protocols for plants–CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer-Verlag, Berlin, pp 283–293
Doyle JA, Le Thomas A (1996) Phylogenetic analysis and character evolution in Annonaceae. Bull Mus Natl Hist Nat, B, Adansonia 3–4:279–334
Doyle JA, Le Thomas A (1997a) Significance of palynology for phylogeny of Annonaceae: experiments with removal of pollen characters. Plant Syst Evol 206:133–159
Doyle JA, Le Thomas A (1997b) Phylogeny and geographic history of Annonaceae. Geogr Phys Quatern 51:252–361
Doyle JA, Bygrave P, Le Thomas A (2000) Implications of molecular data for pollen evolution in Annonaceae. In: Harley MM, Morton CM, Blackmore S (eds) Pollen and spores: morphology and biology. Royal Botanical Gardens, Kew, pp 259–284
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and inter simple sequence repeat (ISSR) markers. Mol Ecol 8:443–451
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fang DQ, Roose ML (1997) Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet 95:408–417
Fischer M, Matthies D (1998) Effects of population size on performance in the rare plant Gentianella germanica. J Ecol 86:195–204
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. Am J Bot 87:783–792
González-Astorga J, Castillo-Campos G (2004) Genetic variability of the narrow endemic tree Antirhea aromatica Castillo-Campos and Lorence (Rubiaceae, Guettardeae) in a Tropical Forest of Mexico. Ann Bot 93:521–528
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman and Hall, New York, pp 281–304
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 6:95–124
Hasebe M, Omori T, Nakazawa M, Sano T, Kato M, Iwatsuki K (1994) rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proc Natl Acad Sci USA 91:5730–5734
Hogbin PM, Peakall R (1999) Evaluation of the conservation of genetic research to the management of endangered plant Zieria prostrata. Conserv Biol 13:514–522
Hollingsworth P, Forrest L, Spouge J et al (2009) Polyalthia oblongifolia voucher WP2A0446: ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. GenBank:GQ248677.1 & maturase K (matK) gene, partial cds; chloroplast. GenBank:GQ248183.1
Lewontin RC (1972) The apportionment of human diversity. Evolution Biol 6:381–398
Manos PS, Steel KP (1997) Phylogenetic analysis of “higher” Hamamelididae based on plastid sequence data. Am J Bot 84:1407–1419
Miller MP (1997) Tools for Population Genetic Analyses (TFPGA), ver. 1.3. A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author. http://www.marksgeneticsoftware.net/_vti _bin/shtml.exe/tfpga.html
Mols JB, Co DLV, Gravendeel B, Chatrou LW, Pirie MD, van der Ham RWJM, van Marle EJ, Keßler PJA (2004a) Morphological character evolution in the miliusoid clade (Annonaceae). In: Mols JB (ed) From Miliusa to Miliuseae to Miliusoid: identifying clades in Asian Annonaceae, Ph.D. Thesis, Leiden University, Leiden, pp 37–75
Mols JB, Gravendeel B, Chatrou LW, Pirie MD, Bygrave PC, Chase MW, Keßler PJA (2004b) Identifying clades in Asian Annonaceae: monophyletic genera in the polyphyletic Miliuseae. Am J Bot 91:590–600
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1977) F-Statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet 41:225–233
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qiu YX, Hong DY, Fu CX, Cameron KM (2004) Genetic variation in the endangered and endemic species Changium smyrnioides (Apiaceae). Biochem Sys Ecol 32:583–596
Qiu YX, Li JH, Liu HL, Chen YY, Fu CX (2006) Population structure and genetic diversity of Dysosma versipellis (Berberidaceae), a rare endemic from China. Biochem Sys Ecol 34:745–752
Ratnayake RMCS, Su YCF, Gunatilleke IAUN, Wijesundara DSA, Saunders RMK (2006) Reproductive biology of two sympatric species of Polyalthia (Annonaceae) in Sri Lanka. II. Breeding systems and population genetic structure. Int J Plant Sci 167:495–502
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, John P, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Saunders RMK, Yuonne CFS, Xue B (2011) Phylogenetic affinities of Polyalthia species (Annonaceae) with columellar-sulcate pollen: enlarging the Madagascan endemic genus Fenerivia. Taxon 60:1407–1416
Schemske DW, Husband BC, Ruckelshaus MH, Goodwillie C, Parker IM, Bishop JG (1994) Evaluation approaches to the conservation of rare and endangered plants. Ecology 75:584–606
Su YC, Smith GJ, Saunders RM (2008) Phylogeny of the basal angiosperm genus Pseuduvaria (Annonaceae) inferred from five chloroplast DNA regions, with interpretation of morphological character evolution. Mol Phylogenet Evol 48:188–206
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Phylogenet Evol 28:2731–2739
van Heusden ECH (1992) Flowers of Annonaceae: Morphology, Classification, and Evolution. Blumea, Suppl 7. Leiden
van Setten AK, Koek-Noorman J (1992) Fruits and seeds of Annonaceae. Morphology and its significance for classification and identification. Bibliotheca Botanica, Heft 142. E. Schweizerbart’sche Verlagsbuchhandlung, Stuttgart
Viswanathan MB, Manikandan U (2001) Polyalthia tirunelveliensis (Annonaceae), a new species from Peninsular India. Kew Bull 56:217–221
Viswanathan MB, Rajasekar C, Sathish Kumar P, Rajesh R (2014) Monoon tirunelveliense voucher BDUT 1402: ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. GenBank:KF887447.1 & maturase K (matK) gene, partial cds; chloroplast. GenBank:KF887448.1. http://www.ncbi.nlm.nih.gov/nuccore/KF887448.1
Wang J (2004) Estimating pairwise relatedness from dominant genetic markers. Mol Ecol 13:3169–3178
Wolfe AD, Xiang QY, Kephart SR (1998) Assessing hybridization in natural populations of Penstemon (Scrophulariaceae) using hypervariable inter simple sequence repeat (ISSR) bands. Mol Ecol 7:1107–1125
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Xiao LQ, Ge XJ, Gong X, Hao G, Zheng SX (2004) ISSR Variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae). Ann Bot 94:133–138
XLSTAT (2013). Addinsoft Inc., Paris, France. http://www.xlstat.com
Xue B, Yvonne CFU, Mols JB, Keßler PJA, Saunders RMK (2011) Further fragmentation of the polypheletic genus Polyalthia (Annonaceae): molecular phylogenetic support for a broader delimitation of Marsypopetalum. Syst Biodivers 9:17–26
Xue B, Su YCF, Thomas DC, Saunders RMK (2012) Pruning the polyphyletic genus Polyalthia (Annonaceae) and resurrecting the genus Monoon. Taxon 61:1021–1039
Yeh FC, Yang RC, Boyle T (1999) POPGENE ver. 1.3.1. Microsoft Windows® - based freeware for population genetic analysis. Available: (www.ualberta.ca/~fyeh/) University of Alberta and the Centre for International Forestry Research, Edmonton
Author contribution statement
M.B. Viswanathan Principal Investigator of the Project entitled, “Studies on Population Ecology and Micropropagation Techniques of Selected Endemic and Threatened Plants of Peninsular India” sponsored by Department of Biotechnology, Government of India, New Delhi, for fund assistance (BT/PR-9368/BCE/08/563/2007 dt.25.06.2008). Major Contributor of the paper. C. Rajasekar: Senior Research Fellow of the Project. Next to first author, he contributed by field work and provided basic data. P. Sathish Kumar: Technical Assistant of the Project. He assisted the Senior Research Fellow in the field work and looked after the financial aspects of the Project.
Acknowledgments
The authors are grateful to the Department of Biotechnology, Government of India, New Delhi, for fund assistance (BT/PR-9368/BCE/08/563/2007 dt.25.06.2008), Dr. K.S. Charak, Adviser and Dr. Onkar N. Tiwari, Scientist ‘C’ for their administrative help, the Chief Wildlife Warden, Principal Chief Conservator of Forests, Chennai, and Chief Conservator of Forests and Field Director, Project Tiger, Tirunelveli, for permission to field work.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Carlson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
468_2014_1122_MOESM1_ESM.tif
UPGMA dendrogram of the seven population of M. tirunelveliense constructed by ISSR and compared to Bayesian admixture proportion (K=7) (TIFF 144 kb)
Rights and permissions
About this article
Cite this article
Viswanathan, M.B., Rajasekar, C. & Sathish Kumar, P. Genetic diversity of steno endemic and critically endangered Monoon tirunelveliense (Annonaceae) from India as revealed by ISSRs. Trees 29, 437–447 (2015). https://doi.org/10.1007/s00468-014-1122-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-014-1122-y