Abstract
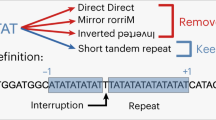
Short tandem repeats (STRs) are subjected to two kinds of mutational modifications: point mutations and replication slippages. The latter is found to be the more frequent cause of STR modifications, but a satisfactory quantitative measure of the ratio of the two processes has yet to be determined. The comparison of entire genome sequences of closely enough related species enables one to obtain sufficient statistics by counting the differences in the STR regions. We analyzed human–chimpanzee DNA sequence alignments to obtain the counts of point mutations and replication slippage modifications. The results were compared with the results of a computer simulation, and the parameters quantifying the replication slippage probability as well as the probabilities of point mutations within the repeats were determined. It was found that within the STRs with repeated units consisting of one, two or three nucleotides, point mutations occur approximately twice as frequently as one would expect on the basis of the 1.2% difference between the human and chimpanzee genomes. As expected, the replication slippage probability is negligible below a 10-bp threshold and grows above this level. The replication slippage events outnumber the point mutations by one or two orders of magnitude, but are still lower by one order of magnitude relative to the mutability of the markers that are used for genotyping purposes.



Similar content being viewed by others
References
Arndt PF, Hwa T, Petrov DA (2005) Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. J Mol Evol 60:748–763
Bell GI (1996) Evolution of simple sequence repeats. Comp Chem 20:41–48
Borštnik B, Pumpernik D (2002) Tandem repeats in protein coding regions of primate genes. Genome Res 12:909–915
Borštnik B, Pumpernik D (2004) Mutational dynamics of short tandem repeats in human genome. Europhys Lett 65:290–296
Borštnik B, Pumpernik D (2005) Evidence on DNA slippage step-length distribution. Phys Rev E 71:031913-1–031913-7
Borštnik B, Pumpernik D, Lukman D (1993) Analysis of apparent 1/f spectrum in DNA sequences. Europhys Lett 23:389–394
Cox R, Mirkin SM (1997) Characteristic enrichment of DNA repeats in different genomes. Proc Natl Acad Sci USA 94:5237–5242
Debrauwere H, Gendrel CG, Lechat S, Dutreix M (1997) Differences and similarities between various tandem repeat sequences: minisatellites and microsatellites. Biochemie 79:577–586
El-Sawy M, Deininger P (2005) Tandem insertions of Alu elements. Cytogenet Genome Res 108:58–62
Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE (2005) Why do human diversity vary at a megabase scale? Genome Res 15:1222–1231
International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 43:1931–1945
Jurka J (2004) Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev 14:603–608
Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ (2003) The UCSC genome browser database. Nucleic Acids Res 31:51–54
Karthikeyan G, Chary KVR, Rao BJ (1999) Fold-back structures at the distal end influence DNA slippage at the proximal end during mononucleotide repeat expansions. Nucleic Acids Res 27:3851–3858
Kayser M, Vowles EJ, Kappei D, Amos W (2006) Microsatellite length differences between humans and chimpanzees at autosomal but not Y-chromosomal loci. Genetics 173:2179–2186
Kondrashov AS (2003) Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 21:12–27
Kruglyak S, Durett RT, Schug MD, Aquadro CF (1998) Equilibrium distribution of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci USA 95:10774–10778
Kumar S, Subramanian S (2002) Mutation rates in mammalian genomes. Proc Natl Acad Sci USA 99:803–808
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Sainudiin R, Durrett RT, Aquadro CF, Nielsen R (2004) Microsatellite mutation models: insight from a comparison from humans and chimpanzees. Genetics 168:383–395
Savage D, Batley J, Erwin T, Logan E, Love CG, Lim GAC, Mongin E, Barker G, Spangenberg GC, Edwards D (2005) SNPServer: a real-time SNP discovery tool. Nucleic Acids Res 33:W493–W495
The Chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87
The International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933
Varki A, Altheide TK (2005) Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res 15:1746–1758
von Gruenberg HH, Peifer M, Timmer J, Kollmann M (2004) Variations in substitution rate in human and mouse genomes. Phys Rev Lett 93:208102-1–208102-4
Vowles EJ, Amos W (2006) Quantifying ascertainment bias and species-specific length differences in human and chimpanzee microsatellites using genome sequences. Mol Biol Evol 23:598–607
Webster MT, Smith NGC, Ellegren H (2002) Microsatellite evolution inferred from human–chimpanzee genomic sequence alignments. Proc Natl Acad Sci USA 99:8748–8753
Yang S, Smit AF, Schwartz S, Chiaromonte F, Roskin KM, Haussler D, Miller W, Hardison RC (2004) Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res 14:17–527
Zhivotovsky LA (2001) Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 18:700–709
Zhivotovsky LA, Feldman MW (1995) Microsatellite variability and genetic distances. Proc Natl Acad Sci USA 92:11549–11552
Acknowledgments
This work was supported by the Slovenian Research Agency. The anonymous referees’ comments are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Van de Peer.
Rights and permissions
About this article
Cite this article
Pumpernik, D., Oblak, B. & Borštnik, B. Replication slippage versus point mutation rates in short tandem repeats of the human genome. Mol Genet Genomics 279, 53–61 (2008). https://doi.org/10.1007/s00438-007-0294-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0294-1