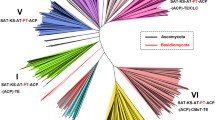
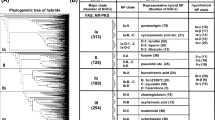
Abstract
Zingiberaceae or ‘ginger family’ is the largest family in the order ‘Zingiberales’ with more than 1300 species in 52 genera, which are mostly distributed throughout Asia, tropical Africa and the native regions of America with their maximum diversity in Southeast Asia. Many of the members are important spice, medicinal or ornamental plants including ginger, turmeric, cardamom and kaempferia. These plants are distinguished for the highly valuable metabolic products, which are synthesised through phenylpropanoid pathway, where type III polyketide synthase is the key enzyme. In our present study, we used sequence, structural and evolutionary approaches to scrutinise the type III polyketide synthase (PKS) repertoire encoded in the Zingiberaceae family. Highly conserved amino acid residues in the sequence alignment and phylogram suggested strong relationships between the type III PKS members of Zingiberaceae. Sequence and structural level investigation of type III PKSs showed a small number of variations in the substrate binding pocket, leading to functional divergence among these PKS members. Molecular evolutionary studies indicate that type III PKSs within Zingiberaceae evolved under strong purifying selection pressure, and positive selections were rarely detected in the family. Structural modelling and protein-small molecule interaction studies on Zingiber officinale PKS ‘a representative from Zingiberaceae’ suggested that the protein is comparatively stable without much disorder and exhibited wide substrate acceptance.













Similar content being viewed by others
References
Abe I, Takahashi Y, Morita H, Noguchi H (2001) Benzalacetone synthase. A novel polyketide synthase that plays a crucial role in the biosynthesis of phenylbutanones in Rheum palmatum. Eur J Biochem 268(11):3354–9
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–33
Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20(1):79–110
Austin MB, Izumikawa M, Bowman ME (2004) Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J Biol Chem 279:45162–74
Brand S, Holscher D, Schierhorn A, Svatos A, Schroder J, Schneider B (2006) A type III polyketide synthase from Wachendorfia thyrsiflora and its role in diarylheptanoid and phenylphenalenone biosynthesis. Planta 224:413–28
Cheng J, Baldi P (2007) Improved residue contact prediction using support vector machines and a large feature set. BMC Bioinforma 8:113
Chizzali C, Beerhues L (2012) Phytoalexins of the pyrinae: biphenyls and dibenzofurans. Beilstein J Org Chem 8:613–20
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Dao TT, Linthorst HJ, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 3:397–412
Edwards ML, Stemerick DM, Sunkara PS (1990) Chalcones: a new class of antimitotic agents. J Med Chem 33:1948–54
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol 6:775–84
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Helariutta Y, Kotilainen M, Elomaa P, Kalkkinen N, Bremer K, Teeri TH, Albert VA (1996) Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proc Natl Acad Sci 93(17):9033–8
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23(2):254–267
Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM (1997) Cancer chemopreventive activity of resveratrol., a natural product derived from grapes. Science 275:218–20
Jez JM, Austin MB, Ferrer J, Bowman ME, Schroder J, Noel JP (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol 7(12):919–30
Jez JM, Bowman ME, Noel JP (2001) Structure-guided programming of polyketide chain-length determination in chalcone synthase. Biochemistry 40(49):14829–38
Jez JM, Bowman ME, Noel JP (2002) Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc Natl Acad Sci U S A 99:5319–24
Jiang H, Timmermann BN, Gang DR (2006) Use of liquid chromatography-electrospray ionization tandem mass spectrometry to identify diarylheptanoids in turmeric (Curcuma longa L.) rhizome. J Chromatogr 1111:21–31
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Katsuyama Y, Kita T, Funa N, Horinouchi S (2009a) Identification and characterization of multiple curcumin synthases from the herb Curcuma longa. FEBS Lett 583:2799–2803
Katsuyama Y, Kita T, Funa N, Horinouchi S (2009b) Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J Biol Chem 284:11160–11170
Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:387–392
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Koes RE, Spelt CE, Mol JNM, Geratas AGM (1987) The chalcone synthase multigene family of Petunia hybrida (V30) Sequence homology, chromosomal location and evolutionary aspects. Plant Mol Biol 10:375–85
Koo HJ, McDowell ET, Ma X, Greer KA, Kapteyn J, Xie Z, Descour A, Kim H, Yu Y, Kudrna D, Wing RA, Soderlund CA, Gang DR (2013) Ginger and turmeric expressed sequence tags identify signature genes for rhizome identity and development and the biosynthesis of curcuminoids., gingerols and terpenoids. BMC Plant Biol 13:27. doi:10.1186/1471-2229-13-27
Koonin EV, Wolf YI (2010) Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet 11(7):487–98
Koskela S, Elomaa P, Helariutta Y, Soderholm P, Vuorela P (2001) Two bioactive compounds and a novel chalcone synthase like enzyme identified in Gerbera hybrida. Proc. IV IS on In Vitro Cult. & Hort Breeding. ISHS Acta Hort 560:271–4
Kumar S, Gadagkar SR (2001) Disparity index: a simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics 158:1321–1327
Lakshmi S, Padmaja G, Remani P (2011) Antitumour Effects of Isocurcumenol Isolated from Curcuma zedoaria Rhizomes on Human and Murine Cancer Cells. Int J Med Chem. doi:10.1155/2011/253962
Lanz T, Tropf S, Marner FJ, Schroder J, Schroder G (1991) The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J Biol Chem 266(15):9971–6
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lee HS, Zhang Y (2012) BSP-SLIM: a blind low-resolution ligand-protein docking approach using theoretically predicted protein structures. Proteins 80:93–110
Li R, Kenyon GL, Cohen FE, Chen X, Gong B, Dominguez JN, Davidson E, Kurzban G, Miller RE, Nuzum EO (1995) In vitro antimalarial activity of chalcones and their derivatives. J Med Chem 38(26):5031–7
Linding R, Russell RB, Neduva V, Gibson TJ (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:13
Lovell SC, Davis IW, Arendall WB, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by Calpha geometry: phi., psi and Cbeta deviation. Proteins: structure. Funct Genet 50:437–450
Lukacin R, Schreiner S, Matern U (2001) Transformation of acridone synthase to chalcone synthase. FEBS Lett 508(3):413–7
Mallika V, Sivakumar KC, Soniya EV (2011) Evolutionary implications and physicochemical analyses of selected proteins of type III polyketide synthase family. Evol Bioinform Online 7:41–53
Mo Y, Nagel C, Taylor LP (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci U S A 89:7213–7217
Morita, H., Abe, I., Noguchi, H (2010) Plant type III PKS. Comprehensive Natural Products II. Lew Mander and Hung-Wen (Ben) Liu (ed) Oxford: 171–225.
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–5
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Okada Y, Ito K (2001) Cloning and analysis of valerophenone synthase gene expressed specifically in lupulin gland of hop (Humulus lupulus L.). Biosci Biotechnol Biochem 65:150–155
Radhakrishnan EK, Sivakumar KC, Soniya EV (2009) Molecular characterization of novel form of type III polyketide synthase from Zingiber Officinale Rosc., its analysis using bioinformatics method. J Proteomics Bioinform 2:310–5
Radhakrishnan EK, Rintu TV, Soniya EV (2010) Unusual intron in the second exon of a type III polyketide synthase gene of Alpinia calcarata Rosc. Genet Mol Biol 33(1):141–145
Resmi MS, Soniya EV (2012) Molecular cloning and differential expressions of two cDNA encoding type III polyketide synthase in different tissues of Curcuma longa L. Gene 491(2):278–83
Resmi MS, Verma P, Gokhale RS, Soniya EV (2013) Identification and characterization of a type III polyketide synthase involved in quinolone alkaloid biosynthesis from Aegle marmelos Correa. J Biol Chem 288(10):7271–81
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:320–324
Rozas J (2009) DNA sequence polymorphism analysis using DnaSP. In: Posada D (ed.) Bioinformatics for DNA Sequence Analysis; Methods in Molecular Biology Series, Humana Press. USA, 537:pp. 337–350
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–25
Sander C, Schneider R (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9(1):56–68
Sanders IF (2008) Sequencing of Medicago truncatula genome and studies of metabolic gene organization and expression profile. PhD diss., The University of Oklahoma, ProQuest,. http://gradworks.umi.com/33/07/3307965.html
Schijlen EG, Ric de Vos CH, Van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65(19):2631–48
Schroder J (1997) A family of plant—specific polyketide synthases: facts and predictions. Trends Plant Sci 2:373–378
Schroder J, Schroder G (1990) Stilbene and chalcone synthases: related enzymes with key functions in plant-specific pathways. Z Naturforsch C 45:1–8
Schroder G, Schroder J (1992) A single change of histidine to glutamine alters the substrate preference of a stilbene synthase. J Biol Chem 267(29):20558–60
Steinway SN, Dannenfelser R, Laucius CD, Hayes JE, Nayak S (2010) JCoDA: a tool for detecting evolutionary selection. BMC Bioinforma 11:284
Stewart CJ, Vickery CR, Burkart MD, Noel JP (2013) Confluence of structural and chemical biology: plant polyketide synthases as biocatalysts for a bio-based future. Curr Opin Plant Biol 16(3):365–72
Suh DY, Fukuma K, Kagami J, Yamazaki Y, Shibuya M, Ebizuka Y, Sankawa U (2000) Identification of amino acid residues important in the cyclization reactions of chalcone and stilbene synthases. Biochem J 350:229–235
Tajima F (1993) Simple methods for testing molecular clock hypothesis. Genetics 135:599–607
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood., evolutionary distance., maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vehlow C, Stehr H, Winkelmann M, Duarte JM, Petzold L, Dinse J, Lappe M (2011) CMView: interactive contact map visualization and analysis. Bioinformatics 27(11):1573–4
Wakimoto T, Morita H, Abe I (2012) Engineering of plant type III polyketide synthases. Methods Enzymol 515:337–58
Wanibuchi K, Zhang P, Abe T (2007) An acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata. FEBS J 274(4):1073–82
Watanabe K, Praseuth AP, Wang CC (2007) A comprehensive and engaging overview of the type III family of polyketide synthases. Curr Opin Chem Biol 11:279–286
Wu S, Zhang Y (2008) A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics 24:924–931
Xing Y, Lee C (2005) Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci U S A 102(38):13526–13531
Xu D, Zhang Y (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80:1715–1735
Yang Z, Nielsen R, Goldman N, Pedersen AM (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155(1):431–49
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64(3):643–51
Zhang J, Yu L, Yang Z (2011) Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19:1784–1795
Zhou L, Wang Y, Peng Z (2011) Molecular characterization and expression analysis of chalcone synthase gene during flower development in tree peony (Paeonia suffruticosa). Afr J Biotechnol 10:1275–1284
Zmasek CM, Eddy SR (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 4:383–4
Acknowledgments
We thank the anonymous reviewers for the useful comments, which helped us to improve the manuscript. The authors are thankful to Council of Scientific and Industrial Research (CSIR) for Senior Research Fellowship (09/716/(0146)/2012/EMR-I) and Department of Science and Technology (DST) for financial support. Authors are also acknowledging Department of Biotechnology (DBT) for the facilities provided.
Author contributions
Conceived and designed the experiments: VM, PTG, AR and EVS. Performed the experiments: VM, PTG and AR. Analysed the data: VM and GA. Wrote the paper: VM, GA and EVS
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Sureshkumar Balasubramanian
P. T. Gincy and A. Remakanthan have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Mallika, V., Aiswarya, G., Gincy, P.T. et al. Type III polyketide synthase repertoire in Zingiberaceae: computational insights into the sequence, structure and evolution. Dev Genes Evol 226, 269–285 (2016). https://doi.org/10.1007/s00427-016-0548-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-016-0548-1