Abstract
Alternative oxidase (AOX) is a diiron carboxylate protein present in all plants examined to date that couples the oxidation of ubiquinol with the reduction of oxygen to water. The predominant structure of AOX genes is four exons interrupted by three introns. In this study, by analyzing the genomic sequences of genes from different plant species, we deduced that intron/exon loss/gain and deletion of fragments are the major mechanisms responsible for the generation and evolution of AOX paralogous genes. Integrating gene duplication and structural information with expression profiles for various AOXs revealed that tandem duplication/block duplication contributed greatly to the generation and maintenance of the AOX gene family. Notably, the expression profiles based on public microarray database showed highly diverse expression patterns among AOX members in different developmental stages and tissues and that both orthologous and paralogous genes did not have the same expression profiles due to their divergence in regulatory regions. Comparative analysis of genes in six plant species under various perturbations indicated a large number of protein kinases, transcription factors and antioxidant enzymes are co-expressed with AOX. Of these, four sets of transcription factors—WRKY, NAC, bZIP and MYB—are likely involved in the regulating the differential responses of AOX1 genes to specific stresses. Furthermore, divergence of AOX1 and AOX2 subfamilies in regulation might be the main reason for their differential stress responses.




Similar content being viewed by others
References
Albury MS, Elliott C, Moore AL (2010) Ubiquinol-binding site in the alternative oxidase: mutagenesis reveals features important for substrate binding and inhibition. Biochim Biophys Acta 1797:1933–9
Allocco DJ, Kohane IS, Butte AJ (2004) Quantifying the relationship between co-expression, co-regulation and gene function. BMC bioinf 5:18
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Atteia A, van Lis R, van Hellemond JJ, Tielens AGM, Martin W, Henze K (2004) Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX). Gene 330:143–148
Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16:1667–78
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Carmel L, Wolf YI, Rogozin IB, Koonin EV (2007) Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res 17:1034–1044
Carré J, Affourtit C, Moore A (2011) Interaction of purified alternative oxidase from thermogenic Arum maculatum with pyruvate. FEBS Lett 585:397
Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58:193–212
Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1757:730–741
Cohen NE, Shen R, Carmel L (2012) The role of reverse transcriptase in intron gain and loss mechanisms. Mol Biol Evol 29:179–86
Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH (2002) Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol 129:949–953
Costa JH, McDonald AE, Arnholdt-Schmitt B, Fernandes de Melo D (2014) A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 19(Pt B):172–83
Crichton PG, Albury MS, Affourtit C, Moore AL (2010) Mutagenesis of the Sauromatum guttatum alternative oxidase reveals features important for oxygen binding and catalysis. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1797:732–737
Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165
Dinant M, Baurain D, Coosemans N, Joris B, Matagne RF (2001) Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Curr Genet 39:101–108
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Fawcett JA, Rouze P, Van de Peer Y (2012) Higher intron loss rate in Arabidopsis thaliana than A. lyrata is consistent with stronger selection for a smaller genome. Mol Biol Evol 29:849–59
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183:557–564
Frederico AM, Zavattieri MA, Campos MD, Cardoso HG, McDonald AE, Arnholdt‐Schmitt B (2009) The gymnosperm Pinus pinea contains both AOX gene subfamilies, AOX1 and AOX2. Physiol Plant 137:566–577
Grant N, Onda Y, Kakizaki Y, Ito K, Watling J, Robinson S (2009) Two cys or not two cys? That is the question; alternative oxidase in the thermogenic plant sacred lotus. Plant Physiol 150:987–995
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series
Hepburn NJ, Schmidt DW, Mower JP (2012) Loss of two introns from the magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol Biol Evol 29:3111–20
Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA (2003) A tomato alternative oxidase protein with altered regulatory properties. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1606:153–162
Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinf 2008:420747
Jeffares DC, Mourier T, Penny D (2006) The biology of intron gain and loss. TRENDS in Genet 22:16–22
Jiang W-k, Liu Y-l, Xia E-h, Gao L-z (2013) Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol 161:1844–1861
Knowles DG, McLysaght A (2006) High rate of recent intron gain and loss in simultaneously duplicated Arabidopsis genes. Mol Biol Evol 23:1548–1557
Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–8
Lynch M (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Lynch M, Kewalramani A (2003) Messenger RNA surveillance and the evolutionary proliferation of introns. Mol Biol Evol 20:563–571
Lynch M, Richardson AO (2002) The evolution of spliceosomal introns. Curr Opin Genet Dev 12:701–710
McDonald AE, Vanlerberghe GC (2006) Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp Biochem Physiol Part D Genomics Proteomics 1:357–64
Meeuse BJD (1975) Thermogenic respiration in aroids. Annu Rev Plant Physiol 26:117–126
Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD (2010) PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the gene ontology consortium. Nucleic Acids Res 38:D204–10
Millar AH, Hoefnagel MHN, Day DA, Wiskich JT (1996) Specificity of the organic acid activation of alternative oxidase in plant mitochondria. Plant Physiol 111:613–618
Millenaar F, Lambers H (2008) The alternative oxidase: in vivo regulation and function. Plant Biol 5:2–15
Mitrovich QM, Tuch BB, Francisco M, Guthrie C, Johnson AD (2010) Evolution of yeast noncoding RNAs reveals an alternative mechanism for widespread intron loss. Science 330:838–841
Moore AL, Albury MS (2008) Further insights into the structure of the alternative oxidase: from plants to parasites. Biochem Soc Trans 36:1022
Moore AL, Shiba T, Young L, Harada S, Kita K, Ito K (2013) Unraveling the heater: new insights into the structure of the alternative oxidase. Annu Rev Plant Biol 64:637–663
Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7, e31944
Mourier T, Jeffares DC (2003) Eukaryotic intron loss. Science 300:1393–1393
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–52
Neimanis K, Staples JF, Huner NP, McDonald AE (2013) Identification, expression, and taxonomic distribution of alternative oxidases in non-angiosperm plants. Gene 526:275–86
Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA (2013a) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288:3449–3459
Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, Walker H, Van Breusegem F, Whelan J, Giraud E (2013b) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25:3450–71
Oliver SN, Lunn JE, Urbanczyk-Wochniak E, Lytovchenko A, Van Dongen JT, Faix B, Schmälzlin E, Fernie AR, Geigenberger P (2008) Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol 148:1640–1654
Polidoros AN, Mylona PV, Arnholdt‐Schmitt B (2009) Aox gene structure, transcript variation and expression in plants. Physiol Plant 137:342–353
Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell Online 21:3718–3731
Pu X, Lv X, Tan T, Fu F, Qin G,Lin H (2015) Roles of mitochondrial energy dissipation systems in plant development and acclimation to stress. Ann Bot: mcv063
Rhoads D, Umbach A, Sweet C, Lennon A, Rauch G,Siedow J (1998) Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine-residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. The Journal of biological chemistry, 273
Rodríguez-Trelles F, Tarrío R, Ayala FJ (2006) Origins and evolution of spliceosomal introns. Annu Rev Genet 40:47–76
Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV (2003) Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13:1512–1517
Rogozin IB, Carmel L, Csuros M,Koonin EV (2012) Origin and evolution of spliceosomal introns. Biol Direct, 7
Roy SW, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7:211–221
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–38
Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20:461–4
Shiba T, Kido Y, Sakamoto K, Inaoka DK, Tsuge C, Tatsumi R, Takahashi G, Balogun EO, Nara T, Aoki T, Honma T, Tanaka A, Inoue M, Matsuoka S, Saimoto H, Moore AL, Harada S, Kita K (2013) Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc Natl Acad Sci U S A 110:4580–5
Umbach AL, Siedow JN (1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103:845–854
Umbach AL, Ng VS, Siedow JN (2006) Regulation of plant alternative oxidase activity: a tale of two cysteines. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1757:135–142
Vanlerberghe GC (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14:6805–6847
Vanlerberghe GC, Cvetkovska M, Wang J (2009) Is the maintenance of homeostatic mitochondrial signaling during stress a physiological role for alternative oxidase? Physiol Plant 137:392–406
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci 109:1187–1192
Yang YF, Zhu T, Niu DK (2013) Association of intron loss with high mutation rate in Arabidopsis: implications for genome size evolution. Genome Biol Evol 5:723–33
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–32
Further Reading
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–70
Prelic A, Bleuler S, Zimmermann P, Wille A, Buhlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22:1122–9
Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W (2008) Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1:851–857
Acknowledgments
We would like to thank Prof. J.C.M. (Sjef) Smeekens and Dr. Alessia Peviani at Utrecht University, for their helpful advice in visualizing phylogenetic tree, and our lab members, Ting-Hong Tan for providing plant picture. This study was supported by the National Natural Science Foundation of China (31470342, 91417305, 31400211), the National Basic Research Program of China (973 Program) (2015CB150100), and Sichuan Natural Science Foundation (2015JY0101, 2015JY0223).
We thank anonymous reviewers for helpful comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Sureshkumar Balasubramanian
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1aFig. S1b
Three-dimensional structure of predicted AOX. Amino acid residues involved in the diiron active site are shown as pink spheres and hydrogen bonds are indicated by white color. Accession numbers for each gene are listed in supplementary table S1. Three-dimensional structure of AOX Proteins were predicted by the I-TASSER server. The figures were prepared with Pymol1.6 (http://www.pymol.org/). (GIF 3063 kb)
The dimerization potential of AOX. (GIF 5774 kb)
Fig. S2
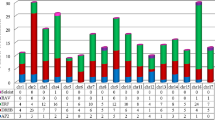
Gene expression profiles of AOX gene family across a diversity of plant species over the development stage. Error bars represent standard error. Data were retrieved from the Genevestigator microarray database (https://www.genevestigator.com/gv/plant.jsp) using development tools. (GIF 3252 kb)
Fig. S3
a-h Expression profiles of the AOX gene family from some plants. The figure represents the average of expression value of log2-scale from diverse tissues or organs of plants and displayed as nodes of a tree which children nodes are included into parent nodes that reflect number of the samples. Error bars indicated standard error. Data were obtained from the Genevestigator microarray database using anatomy tools. (GIF 826 kb)
Fig. S4
A-D Heat map of co-expressed genes. The co-expression analysis was conducted by using Genevestigator co-expression tools with the parameters of 2 log2 fold change and p ≤ 0.05 was used as cut-off. The maximum number of genes for each co-expressed gene lists is 100. (Zimmermann et al., 2008; Hruz et al., 2008). For GO annotations, the co-expressed gene lists generated from co-expression tools were loaded into agriGO (Du et al., 2010) for further analysis with suggested parameters. Hierarchical clustering was conducted by Genevestigator-hierarchical clustering tools using Euclidean distance (Prelic et al., 2006). (GIF 4526 kb)
(GIF 1771 kb)
(GIF 2145 kb)
(GIF 24884 kb)
Fig. S5a-b
Multiple alignment of AOX and PTOX (GIF 4307 kb)
(GIF 4361 kb)
Fig. S6
GO annotations. (DOC 1086 kb)
Table S1
Original sequence identifiers for plant’s AOX gene. (XLS 40 kb)
Table S2
Putative cis-acting elements of AOX gene family member (DOC 119 kb)
Table S3a
Co-expression profiles of AOX-positive-correlation. (XLS 431 kb)
Table S3b
Co-expression profiles of AOX-negative-correlation. (XLS 531 kb)
Rights and permissions
About this article
Cite this article
Pu, Xj., Lv, X. & Lin, Hh. Unraveling the evolution and regulation of the alternative oxidase gene family in plants. Dev Genes Evol 225, 331–339 (2015). https://doi.org/10.1007/s00427-015-0515-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-015-0515-2