Abstract
Many aspects of human physiology and behavior display rhythmicity with a period of approximately 24 h. Rhythmic changes are controlled by an endogenous time keeper, the circadian clock, and include sleep-wake cycles, physical and mental performance capability, blood pressure, and body temperature. Consequently, many diseases, such as metabolic, sleep, autoimmune and mental disorders and cancer, are connected to the circadian rhythm. The development of therapies that take circadian biology into account is thus a promising strategy to improve treatments of diverse disorders, ranging from allergic syndromes to cancer. Circadian alteration of body functions and behavior are, at the molecular level, controlled and mediated by widespread changes in gene expression that happen in anticipation of predictably changing requirements during the day. At the core of the molecular clockwork is a well-studied transcription-translation negative feedback loop. However, evidence is emerging that additional post-transcriptional, RNA-based mechanisms are required to maintain proper clock function. Here, we will discuss recent work implicating regulated mRNA stability, translation and alternative splicing in the control of the mammalian circadian clock, and its role in health and disease.
Similar content being viewed by others
Basic principles of circadian biology
The daily cycle of night and day, leading to oscillating changes in light-dark and temperature conditions, influences all aspects of life. This is true for animals in the wild but also for human beings, who, despite being independent of the sun for light and heat, still follow a 24-h rhythm. The predictable daily repetition of sleep-wake cycles, activity and rest phases, food intake and digestion, etc. allows the body to anticipate requirements and prepare for upcoming tasks. Therefore, many physiological and behavioral aspects are controlled in a periodic, daily manner. For example, blood pressure and digestive activity increase in the morning whereas the production of melatonin is increased in the evening. This occurs in anticipation of activity and food intake or sleep, respectively. The internal system that produces such a 24-h rhythm is called the circadian clock, a cell autonomous oscillator with a period of around 24 h that is present in nearly every cell of the body. In mammals, the tight connection and crosstalk between individual cells of the same tissue lead to the formation of interconnected, organ-specific clocks. This ensures that cells of the same organ are in the same phase and in addition, the oscillation of a (large) group of connected cells is more robust than the oscillation of a single cell alone [21]. The whole system is controlled by, synchronized and aligned with the environment through signals emanating from the suprachiasmatic nucleus (SCN), a region in the brain referred to as the central or master clock [41]. The SCN itself receives direct light input from the eye, which is the most important external cue to synchronize the circadian clock [13]. In the SCN, light activates signaling cascades resulting in neuronal or humoral output, which transmit the signal to other cells and organs collectively called peripheral clocks (Fig. 1). The exposure to light thus acts as a daily resetting signal for the SCN, which then synchronizes peripheral clocks accordingly. In changing external conditions, e.g., upon traveling to another time zone, the body can readjust to the new light-dark phase, which then allows an adaptation of physiological functions to the new environment. As the circadian clock is relatively stable against altered extracellular conditions, this adjustment requires approximately 1 day per hour of phase shift. Importantly, physiological and behavioral cycles with a period of approximately 24 h are maintained in the absence of external cues showing that the circadian clock is a self-sustained system producing a stable oscillation. The features described above, a period of approximately 24 h, entrainability to changing external conditions and the persistence in the absence of external cues are three criteria that need to be fulfilled for an event to be circadian. A fourth criterion is temperature compensation, meaning that a circadian event is stable over a wide temperature range.
Hierarchical organization of the circadian clock. The master pacemaker of the body is located in the suprachiasmatic nucleus (SCN) which receives direct light input through the eye. Signals from the SCN then synchronize local clocks in the peripheral organs to one another and to the environment. Therefore, peripheral clocks show about 4-h phase delay with respect to the SCN [45]. Malfunctioning of the circadian clock (top) is associated with diverse diseases in the peripheral organs (bottom)
Circadian clocks are not restricted to mammals or eukaryotes; instead, corresponding mechanisms, some depending on the redox state and not on a transcription-translation feedback loop (Fig. 2 and see below), are present in all three domains of life [14]. This suggests that the presence of a circadian clock, allowing the anticipation of daily rhythms, confers an evolutionary advantage to unicellular as well as multicellular organisms that justifies the production and maintenance of an elaborate clockwork.
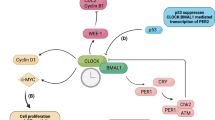
The mammalian circadian clock consists of negative and positive transcriptional feedback loops. At the core of the circadian clock, the BMAL1/CLOCK heterodimer drives oscillating expression of clock-controlled genes (ccgs) with E-box containing promoters. Another layer of regulation is achieved through a second group of ccgs—RORS and REV-ERBs—that regulate BMAL1 transcription. The clock output, such as altered metabolism, is achieved through many genes with E-box containing promoters collectively shown as ccgs (gray). See main text for further details and abbreviations
The circadian clock in health and disease
In humans, many fundamental parameters of the body such as heart rate, blood pressure, body temperature, hormone and cytokine secretion, metabolism, and cerebral activity, show a circadian profile, pointing to a strong influence of the circadian clock on health and disease [62]. The connection and its importance become already apparent when traveling to a different time zone. This leads to a misalignment of the circadian clock with the environment and can cause discomfort and drowsiness, the typical jet lag syndromes. Similarly, “social jet lag”, e.g. the sudden change of daily rhythms at weekends, can disturb the circadian clock and may contribute to the “Monday morning blues” [58]. These examples show that already a temporary disturbance or misalignment of the circadian clock with the environment is sufficient to cause physical and mental uneasiness. In an experimental setting, chronic jet lag increased the mortality rate of old mice [11], further underlining the connection between the circadian system and well-being. Not surprisingly, continued disruption of the circadian rhythm either by genetic or by environmental factors is associated with a variety of diseases in humans (Fig. 1). Especially prominent are cardiovascular diseases, inflammation, cancer, mental health and metabolic disorders, such as diabetes or obesity. The range of diseases may partially reflect the prominent role of the circadian clock in controlling the cell cycle and metabolism [3]. The connection between the circadian clock and pathological conditions has been made and confirmed in different settings such as model organisms, human genetics, and their increased occurrence in shift workers. As an example, long-term shift work has been suggested to increase the risk for colorectal, prostate, and breast cancer as well as cardiovascular and metabolic disorders [57]. Such connections were also found in mice with a genetically disrupted or disturbed clockwork, as these mice show increased incidence of cancer, insulin resistance, and obesity for example [53]. The latter connection is further underlined by the correlation of the time of food intake with weight gain and the development of obesity [2].
The manifestation of several other diseases does not show a direct connection with the circadian system but the severity of their symptoms follows a circadian pattern. Examples include asthma, allergies, or epileptic seizures that preferentially happen at particular times of the day [35, 43], probably associated with circadian histamine and cortisol levels in the blood [7]. Consequently, current efforts are directed toward integrating circadian biology into treatment schedules. If symptoms follow a circadian rhythm, the treatment schedule could be adjusted accordingly; in addition, the body may show very different reactions to the same dose of a drug administered in the morning or the afternoon [6]. First studies have included this aspect in treatment regimens and promising results have been obtained, e.g., in a low-dose prednisone chronotherapy in rheumatoid arthritis patients [8]. Another very active field of research is cancer chronotherapy, as the efficacy as well as the tolerability of chemotherapy shows a circadian profile [44]. While these approaches have a huge potential to improve therapies when applied in general, the future goal of chronomedicine is a personalized treatment schedule based on individual circadian parameters. This will further increase the effectiveness of chronomedicine, as individuals have different chronotypes and the phase and amplitude of the circadian clock changes with aging [50].
In addition to the examples described above, the circadian clock is also connected to neurological and mental disorders. Circadian rhythms play an obvious role in controlling sleep cycles and have been linked to seasonal affective disorder (SAD), a mood disorder with symptoms of depression only during a certain time of the year, depression and neurodegenerative diseases [1, 55, 56]. Another prominent example is the familial advanced sleep phase syndrome, an inheritable disease caused by a point mutation in the PER2 gene. The PER2 S662G mutation destroys a phosphorylation site and thereby changes the stability of the protein, resulting in an altered period and disturbed sleep-wake cycles in the affected individuals [54].
Together, these examples show the strong influence of circadian biology on health and diseases. An extreme example to further illustrate the connection is the dramatically altered mortality of mice when challenged with the same dose of LPS at different times of the day [18], which may be partly explained by micro RNA (miRNA)-based regulation of BMAL1 (see below). In humans, even birth and death show higher occurrence at particular circadian times, with more babies being born in early morning hours [25] and fatalities caused by cardiovascular events peaking between morning and noon [42], emphasizing the central role of the circadian clock in fundamental aspects of life.
Molecular principles of the circadian clock
Rhythmic changes in behavior and physiology are controlled and mediated by extensive circadian changes in gene expression. At the center of the mammalian molecular clock (Fig. 2) is the heterodimeric transcription factor CLOCK/BMAL1 that binds to E-Box containing promoters to activate transcription of up to 10 % of expressed genes (clock-controlled genes, ccgs; [49]). Among the ccgs are the period (PER) and cryptochrome (CRY) genes, that, upon transcription and translation, accumulate in the cytoplasm. Here, the stability of PER and CRY proteins is regulated by ubiquitination—mediated by the E3-ubiqutin ligases βTRCP1 and FBXL21, respectively—and proteasomal degradation [20, 48, 51, 61]. PER/CRY dimers become phosphorylated by the CK1δ/ε kinase and translocate into the nucleus when sufficient amounts of protein have accumulated. In the nucleus, PER/CRY heterodimers inhibit transcription through CLOCK/BMAL1 thus shutting down the expression of ccgs, including themselves; this starts a new cycle once the PER/CRY proteins are degraded and CLOCK/BMAL1 activity is restored [29]. In the nucleus, the stability of PER and CRY proteins is also regulated by ubiquitination. Here, the E3-ubiqutin ligases FBXL3 promotes ubiquitination and proteasomal degradation of CRYs [51]; this activity is antagonized by FBXL21 [20, 61]. A further feedback mechanism is provided through another family of ccgs, RORs and REV-ERBs, which control the transcription of BMAL1 [17]. RORs and REV-ERBs are nuclear receptors that bind to a so-called REV-ERBs/ROR-binding element (RRE) in the promoter region of BMAL1 and regulate its transcription antagonistically. Together, a negative feedback loop is established, which operates with a period of approximately 24 h, and forms the basis for autonomous clocks that are found in a wide variety of cell types in mammalian organisms. In addition to this well-studied transcription-translation feedback loop, post-transcriptional mechanisms have gained increasing attention in the last years, as they were shown to be indispensable for the regulation of the circadian clock. Here, we will focus on recently described post-transcriptional, RNA-based mechanisms in mammalian systems and their impact on circadian biology. For posttranslational modifications and their central role in controlling circadian rhythms, we refer to several recent reviews [29, 39, 52] and references therein.
Emerging RNA-based post-transcriptional mechanisms controlling the mammalian circadian clock
Until recently, E-Box-mediated transcription and its inhibition in a negative transcription-translation feedback loop were believed to be the main rhythmicity inducing mechanism. However, results obtained in the last years indicate that less than 30 % of circadian messenger RNAs (mRNAs) are regulated by de novo transcription, suggesting that other mechanisms account for the majority of circadian gene expression [30, 40]. Indeed, several recent reports confirmed that many, if not all, steps in the life cycle of a (pre-) mRNA can be regulated in a circadian manner thus contributing to circadian gene expression. Core nuclear processing events of mammalian pre-mRNAs are capping, splicing, polyadenylation, and export to the cytoplasm, which will be discussed in the context of circadian biology in the following.
Processing of mammalian pre-mRNA starts with the modification of its 5’ end, the formation of the cap-structure. Regulation of capping has not been documented to be under circadian control, but this may simply reflect that is was not systematically analyzed in circadian settings. It was indeed suggested, that knock down of the RNMT methylase required for cap formation prolongs the circadian period [15], which could indicate that cap formation and its effect on mRNA stability or translation are involved in controlling the circadian cycle. Consistent with this idea, two recent reports have documented that translation through the cap-binding protein eIF4E is under circadian, phosphorylation-dependent control (Fig. 3a, b). Upon rhythmic phosphorylation by the mTOR-effector kinase, ribosomal S6 protein kinase 1 (S6K1), the core circadian transcription factor BMAL1 associates with the translation machinery in the cytoplasm and globally promotes protein synthesis. Phosphorylated BMAL1 interacts with the translation machinery through the cap-binding protein complex eIF4E/A/G and the polyA-binding protein thus controlling cap-dependent translation in a circadian manner [34]. The precise role of BMAL1 in this regulation remains unknown, but a model is conceivable, in which phosphorylated BMAL1 provides a scaffold for more efficient assembly of the translational machinery including polyA-binding protein to generally increase cap-dependent translation. It is interesting to note that PolII-mediated transcription has also been proposed to be globally controlled in a circadian manner. RNA-sequencing of liver mRNAs revealed a peak of intron-containing pre-mRNAs at ∼CT15 (circadian time), which might be explained by a general activation of transcription shortly before [30]. Together, these data suggest that in addition to the specific regulation of individual genes, several steps in global gene expression are under control of the circadian clock.
Post-transcriptional mechanisms acting on the (pre-) mRNA to control the mammalian circadian clock. Before an mRNA becomes translated, several highly regulated processing steps have to occur. This includes capping at the 5’ end, splicing, and polyadenylation at the 3’ end. Furthermore, translation and degradation of the mature mRNA are regulated processes as well. The figure summarizes post-transcriptional events in the life cycle of a (pre-) mRNA that have been reported to directly influence the circadian clock and/or to be controlled in a circadian manner. See main text for further details
Translation efficiency can also be directly modulated through phosphorylation of the eIF4E protein [9]. Here, a light- and circadian clock-regulated MAPK/MNK pathway phosphorylates eIF4E to specifically enhance translation of PER1 and PER2 mRNAs. This effect was specific for PER1 and PER2 mRNAs, likely depending on the sequence of the respective 5’ untranslated regions (UTRs), but the nature of the required cis-acting element and potential coregulated mRNAs have not been defined. The widespread use of regulated translation to generate circadian gene expression was further confirmed in two recent large-scale ribosome-profiling approaches. These studies identified cycling transcripts that are additionally controlled at the level of translation but also found hundreds of non-cycling mRNAs displaying rhythmic ribosome footprints [23, 24]. These results suggest a prominent role for regulated efficiency of translation to control circadian gene expression. One example for RNA-based translational control are short upstream open reading frames (uORFs) that can repress translation of the primary ORF [38]. Such uORFs were identified in core clock components such as BMAL1 and CLOCK, which may contribute to the regulation of the circadian clock. Whether these particular uORFs are functionally important remains to be seen; the finding that knock down of a protein required for reinitiation of translation downstream of uORFs leads to a shortened period provides evidence that at least some uORFs, whose precise nature remains to be identified, are involved in circadian control [24].
Translation of PER1 is further modulated by an internal ribosomal entry site (IRES) in the 5’ UTR of its mRNA (Fig. 3c). IRES-mediated translation allows direct recruitment of the 40S ribosome subunit to the vicinity of the initiation codon and is therefore independent of the classical cap-dependent translation machinery. The IRES element of PER1 was sufficient for rhythmic protein expression of a bicistronic luciferase reporter in synchronized cells thus providing another means to control PER1 expression post-transcriptionally. IRES-mediated translation required rhythmic interaction of the PER1 IRES element with the heterogeneous nuclear ribonucleoprotein Q (hnRNP Q), which activates non-canonical PER1 translation in a circadian manner [33]. The importance of IRES-mediated translation was further confirmed by mathematical modeling and might be regulated through rhythmic phosphorylation of hnRNP Q [32]. Similarly, hnRNP Q and polypyrimidine tract-binding protein (PTB) have been shown to modulate IRES-mediated translation of Reverbα [28].
Another major RNA-processing event, alternative splicing, has been found to be regulated in a circadian manner and to control different aspects of the circadian clock. In general, alternative splicing is a widespread mechanism that can control gene expression in response to changing extracellular conditions [19]. Systematic evidence for mammalian circadian alternative splicing came only recently from a microarray-based approach that suggested many exons to be alternatively spliced in a circadian manner in mouse liver. Furthermore, this microarray suggested circadian mRNA expression of 62 RNA-binding proteins, which might be involved in the regulation of circadian RNA-processing events [37]. The first functional connection between alternative splicing and the mammalian circadian clock was described for the U2AF26 gene that shows circadian and light-induced skipping of exons 6 and 7 in mouse cerebellum (Fig. 3d). This exon-skipping event does not result in a shorter protein product, but, through inducing a frameshift, allows translation into the supposed 3’ UTR adding a new C-terminus to the protein. Interestingly, this new domain shows homology to parts of a drosophila protein called Timeless, which is a central component of the fly circadian clock. This novel U2AF26 isoform localizes to the cytoplasm and, in analogy to Timeless, interacts with PER1 to control its stability. Furthermore, U2AF26-deficient mice show defects in the core molecular clockwork and clock resetting under jet lag conditions, providing evidence for a functional role of U2AF26 alternative splicing in controlling the circadian clock [47]. Another functionally interesting case of alternative isoform expression has been described for melanopsin (Opn4). Here, different isoforms generated by coupled alternative splicing and polyadenylation mediate different responses to light [22]; the regulation of these alternative splicing events and a potential rhythmic production of these isoforms are interesting question to be addressed in future work. Two of the core components of the mammalian circadian clock, CRY1 and PER2, have also been suggested to be alternatively spliced in a circadian manner [4, 46]. However, whether these splicing events are functionally and physiologically relevant remains to be shown.
Further processing events of an mRNA are the addition of a polyA tail, mRNA export into the cytoplasm and the control of mRNA stability, frequently regulated through sequences located in the 3’ UTR. The general efficiency of nuclear mRNA processing and export has been reported to be enhanced through m6A methylation, both in a global manner and specifically for certain core clock transcripts (Fig. 3e; [15]). Consistent with a role of m6A methylation in the circadian cycle and mRNA export, knock down of the methylase Mettl3 elongated the circadian period and led to a general defect in nuclear export of mRNA. While knock down studies and pharmacological inhibition of RNA methylation provide evidence for this regulation, it remains to be shown whether Mettl3 activity and RNA m6A methylation patterns are indeed controlled in a circadian manner.
The 3’ UTR contains many regulatory regions that influence gene expression post-transcriptionally, with important implications for circadian biology. An example are RNA-binding proteins that have been implicated in the control of the circadian clock through controlling mRNA stability of clock components. The hnRNPs K and D both interact with the 3’ UTR of PER3 and antagonistically affect its mRNA stability (Fig. 3f). While hnRNP K stabilizes PER3 mRNA, hnRNP D destabilizes and decreases the amplitude of PER3 mRNA expression [26]. In a similar manner, oscillating cytoplasmatic levels of hnRNP D have been implicated in the modulation of CRY1 mRNA stability [59]. While rhythmic protein expression has been shown or suggested for some hnRNPs such as hnRNP Q [27], hnRNP D [59] and a U2AF26 variant (see above, [47]) this remains to be shown for other RNA-binding proteins. Given the large percentage of circadian mRNAs that are not regulated by de novo transcription, these cases of regulated mRNA stability may only be examples of a widely used mechanism. RNA-binding proteins could selectively stabilize/destabilize target mRNAs at a certain time of the day, thereby yielding a circadian expression profile without changing transcription rates. While the presence of such a regulatory mechanism appears likely, a systematic evaluation of circadian mRNA stability has yet to be performed.
A similarly but equally speculative mechanism would be the presence of circadian miRNAs that could regulate stability or translation of several or many target mRNAs. As a proof of principle it has been shown that miRNA155 can directly target the 3’ UTR of the BMAL1 mRNA to suppress its expression (Fig. 3g). In this case, expression of miRNA155 and its control of BMAL1 expression has been suggested to regulate the different outcomes of the macrophage-mediated innate immunity when stimulated with LPS at different times of the day. Upon macrophage-specific deletion of BMAL1, mice loose the dependence of the mortality rate on the time of the day upon LPS challenge, providing evidence for the involvement of BMAL1 in this phenomenon [10]; however, to confirm a role of miRNA155 in this pathway, such experiments will have to be repeated in miRNA155 deficient animals.
The length of the 3’ UTR can be modulated through alternative polyadenylation, which has recently been linked to the circadian clock. Two cold-inducible RNA-binding proteins have been reported to regulate the choice of alternative polyadenylation sites in the 3’ UTR of clock-controlled genes and may be involved in synchronization of peripheral clocks in response to changing body temperature [36]. Upregulation of Cirbp and Rbm3, as observed under cold-shock, repressed the usage of proximal polyadenylation signals, resulting in prolonged 3’ UTRs of interacting target genes (Fig. 3h). Extended 3’ UTRs may harbor more miRNA-binding sites than the shorter versions [12], which could be associated with a decrease in translation, but further work is needed to confirm such a model in circadian settings. mRNA stability and translation efficiency are furthermore regulated by the length of the polyA tail. Deadenylases are believed to specifically deadenylate certain target mRNAs to yield a selective destabilization. The deadenylase Nocturnin (Noc) is expressed in a circadian manner (Fig. 3i; [5]) and is thus an interesting target to follow this connection. Although Nocturnin knockout mice show resistance to diet-induced obesity and show altered polyA length in a variety of mRNAs [16], these transcripts did not display a circadian change in abundance or polyA length [31]. The potential role of Nocturnin in circadian gene expression and metabolism thus remains to be clarified.
In summary, increasing evidence supports a strong connection of post-transcriptional regulatory events with the mammalian circadian clock. However, investigating the full impact of individual mechanisms such as regulated alternative splicing, mRNA stability or polyadenylation on circadian biology remains a challenging task for the future. In all these cases, global analysis, mechanistic details, and the functional impact of individual components of a group of coregulated (pre-) mRNAs promise exciting discoveries in the coming years.
Conclusions and perspectives
The circadian clock impacts on diverse aspects of human physiology, as has been well-established through elaborate research in the past. Disruption of core clock gene oscillation, either through genetic mutations within these genes or through environmental conditions that interfere with the circadian rhythm, is associated with a variety of diseases. Consequently, chronomedicine is increasingly acknowledged to have a huge potential in improving treatment regimens. Although therapies based on individual chronotypes are still some way in the future, the insights gained through basic research will finally enable the implementation of personalized therapy schedules.
A well-characterized inheritable disease affecting the circadian clock is familial advanced sleep phase syndrome, which is associated with mutations in the CK1δ gene or a point mutation in the PER2 gene which prevents CK1-mediated phosphorylation [54, 60]. This is one example highlighting the importance of posttranslational modifications of core clock genes for circadian biology. In fact, work from the last years conclusively showed that in addition to the core transcription-translational feedback loop, phosphorylation, ubiquitination, and controlled protein degradation are indispensable for proper clock function [29, 39, 52]. Interestingly, circadian control has been investigated mainly at the transcriptional or the posttranslational level, and RNA-based mechanisms contributing to this regulation are only beginning to emerge. Therefore, the contribution of regulated mRNA processing remains exemplary, and a global impact on circadian biology remains to be confirmed. However, given the finding that the majority of circadian mRNAs is not regulated by de novo transcription [30, 40] and the evidence from the studies presented here, we predict that some, if not all, of the mechanisms discussed above will be shown to play prominent and widespread roles in circadian biology. We also consider it likely that after the identification of trans-acting factors involved in RNA-based control of the circadian clock, connections to diseases will follow. A better understanding of the molecular mechanisms that govern the mammalian circadian clock is thus required to first identify such connections, which may then present opportunities for new therapeutic concepts.
References
Albrecht U (2013) Circadian clocks and mood-related behaviors. Handb Exp Pharmacol 227–239. doi:10.1007/978-3-642-25950-0_9
Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW (2009) Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring, Md) 17:2100–2102. doi:10.1038/oby.2009.264
Asher G, Schibler U (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13:125–137. doi:10.1016/j.cmet.2011.01.006
Avitabile D, Genovese L, Ponti D, Ranieri D, Raffa S, Calogero A, Torrisi MR (2014) Nucleolar localization and circadian regulation of Per2S, a novel splicing variant of the Period 2 gene. Cellular and molecular life sciences: CMLS 2013/11/10 edn, City, pp 2547–2559
Baggs JE, Green CB (2003) Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol CB 13:189–198
Baraldo M, Furlanut M (2006) Chronopharmacokinetics of ciclosporin and tacrolimus. Clin Pharmacokinet 45:775–788. doi:10.2165/00003088-200645080-00002
Barnes P, FitzGerald G, Brown M, Dollery C (1980) Nocturnal asthma and changes in circulating epinephrine, histamine, and cortisol. N Engl J Med 303:263–267. doi:10.1056/nejm198007313030506
Buttgereit F, Mehta D, Kirwan J, Szechinski J, Boers M, Alten RE, Supronik J, Szombati I, Romer U, Witte S et al (2013) Low-dose prednisone chronotherapy for rheumatoid arthritis: a randomised clinical trial (CAPRA-2). Ann Rheum Dis 72:204–210. doi:10.1136/annrheumdis-2011-201067
Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC et al (2015) Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci 18:855–862. doi:10.1038/nn.4010
Curtis AM, Fagundes CT, Yang G, Palsson-McDermott EM, Wochal P, McGettrick AF, Foley NH, Early JO, Chen L, Zhang H et al (2015) Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A 112:7231–7236. doi:10.1073/pnas.1501327112
Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD (2006) Chronic jet-lag increases mortality in aged mice. Curr Biol CB 16:R914–R916. doi:10.1016/j.cub.2006.09.058
Di Giammartino Dafne C, Nishida K, Manley James L (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43:853–866. doi:10.1016/j.molcel.2011.08.017
Dibner C, Schibler U, Albrecht U (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72:517–549. doi:10.1146/annurev-physiol-021909-135821
Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA et al (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–464. doi:10.1038/nature11088
Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I et al (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155:793–806. doi:10.1016/j.cell.2013.10.026
Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC (2007) Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A 104:9888–9893. doi:10.1073/pnas.0702448104
Guillaumond F, Dardente H, Giguere V, Cermakian N (2005) Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythm 20:391–403. doi:10.1177/0748730405277232
Halberg F, Johnson EA, Brown BW, Bittner JJ (1960) Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med Soc Exp Biol Med (New York, NY) 103:142–144
Heyd F, Lynch KW (2011) Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem Sci 36:397–404. doi:10.1016/j.tibs.2011.04.003
Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152:1106–1118. doi:10.1016/j.cell.2013.01.054
Hogenesch JB, Herzog ED (2011) Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett 585:1427–1434. doi:10.1016/j.febslet.2011.04.048
Jagannath A, Hughes S, Abdelgany A, Pothecary CA, Di Pretoro S, Pires SS, Vachtsevanos A, Pilorz V, Brown LA, Hossbach M et al (2015) Isoforms of melanopsin mediate different behavioral responses to light. Curr Biol CB 25:2430–2434. doi:10.1016/j.cub.2015.07.071
Jang C, Lahens NF, Hogenesch JB, Sehgal A (2015) Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res 25:1836–1847. doi:10.1101/gr.191296.115
Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D (2015) Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res 25:1848–1859. doi:10.1101/gr.195404.115
Kaiser IH, Halberg F (1962) Circadian periodic aspects of birth. Ann N Y Acad Sci 98:1056–1068
Kim SH, Lee KH, Kim DY, Kwak E, Kim S, Kim KT (2015) Rhythmic control of mRNA stability modulates circadian amplitude of mouse Period3 mRNA. J Neurochem 132:642–656. doi:10.1111/jnc.13027
Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT (2007) Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev 21:797–810. doi:10.1101/gad.1519507
Kim DY, Woo KC, Lee KH, Kim TD, Kim KT (2010) hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res 38:7068–7078. doi:10.1093/nar/gkq569
Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15(Spec No 2):R271–R277. doi:10.1093/hmg/ddl207
Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (New York, NY) 338:349–354. doi:10.1126/science.1226339
Kojima S, Gendreau KL, Sher-Chen EL, Gao P, Green CB (2015) Changes in poly(A) tail length dynamics from the loss of the circadian deadenylase Nocturnin. Sci Reports 5:17059. doi:10.1038/srep17059
Lee KH, Kim SH, Kim DY, Kim S, Kim KT (2012) Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PLoS One 7:e37936. doi:10.1371/journal.pone.0037936
Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT (2012) Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol 32:717–728. doi:10.1128/mcb.06177-11
Lipton Jonathan O, Yuan Elizabeth D, Boyle Lara M, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, Asara John M, Sahin M (2015) The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161:1138–1151. doi:10.1016/j.cell.2015.04.002
Litinski M, Scheer FA, Shea SA (2009) Influence of the circadian system on disease severity. Sleep Med Clin 4:143–163. doi:10.1016/j.jsmc.2009.02.005
Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J (2013) Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Reports 3:2054. doi:10.1038/srep02054
McGlincy NJ, Valomon A, Chesham JE, Maywood ES, Hastings MH, Ule J (2012) Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol 13:R54. doi:10.1186/gb-2012-13-6-r54
Medenbach J, Seiler M, Hentze MW (2011) Translational control via protein-regulated upstream open reading frames. Cell 145:902–913. doi:10.1016/j.cell.2011.05.005
Mehra A, Baker CL, Loros JJ, Dunlap JC (2009) Post-translational modifications in circadian rhythms. Trends Biochem Sci 34:483–490. doi:10.1016/j.tibs.2009.06.006
Menet JS, Rodriguez J, Abruzzi KC, Rosbash M (2012) Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1:e00011. doi:10.7554/eLife.00011
Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445–462. doi:10.1146/annurev-neuro-060909-153128
Morris CJ, Yang JN, Scheer FAJL (2012) The impact of the circadian timing system on cardiovascular and metabolic function. Prog Brain Res 199:337–358. doi:10.1016/B978-0-444-59427-3.00019-8
Orihara K, Saito H (2015) Controlling the peripheral clock might be a new treatment strategy in allergy and immunology. J Allergy Clin Immunol. doi:10.1016/j.jaci.2015.08.054
Ortiz-Tudela E, Mteyrek A, Ballesta A, Innominato PF, Levi F (2013) Cancer chronotherapeutics: experimental, theoretical, and clinical aspects. Handb Exp Pharmacol 261–288. doi:10.1007/978-3-642-25950-0_11
Peirson SN, Butler JN, Duffield GE, Takher S, Sharma P, Foster RG (2006) Comparison of clock gene expression in SCN, retina, heart, and liver of mice. Biochem Biophys Res Commun 351:800–807. doi:10.1016/j.bbrc.2006.10.118
Pembroke WG, Babbs A, Davies K, Ponting CP, Oliver PL (2015) Temporal transcriptomics suggest that twin-peaking genes reset the clock. eLife 4. doi:10.7554/eLife.10518
Preussner M, Wilhelmi I, Schultz AS, Finkernagel F, Michel M, Moroy T, Heyd F (2014) Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol Cell 54:651–662. doi:10.1016/j.molcel.2014.04.015
Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A (2007) Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythm 22:375–386. doi:10.1177/0748730407303926
Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9:e1000595. doi:10.1371/journal.pbio.1000595
Roenneberg T, Kuehnle T, Pramstaller PP, Ricken J, Havel M, Guth A, Merrow M (2004) A marker for the end of adolescence. Curr Biol CB 14:R1038–R1039. doi:10.1016/j.cub.2004.11.039
Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS (2007) Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129:1011–1023. doi:10.1016/j.cell.2007.04.030
Stojkovic K, Wing SS, Cermakian N (2014) A central role for ubiquitination within a circadian clock protein modification code. Front Mol Neurosci 7:69. doi:10.3389/fnmol.2014.00069
Takahashi JS, Hong HK, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9:764–775. doi:10.1038/nrg2430
Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science (New York, NY) 291:1040–1043
Videnovic A, Lazar AS, Barker RA, Overeem S (2014) ‘The clocks that time us’—circadian rhythms in neurodegenerative disorders. Nat Rev Neurol 10:683–693. doi:10.1038/nrneurol.2014.206
Waddington Lamont E, Legault-Coutu D, Cermakian N, Boivin DB (2007) The role of circadian clock genes in mental disorders. Dialogues Clin Neurosci 9:333–342
Wang XS, Armstrong MEG, Cairns BJ, Key TJ, Travis RC (2011) Shift work and chronic disease: the epidemiological evidence. Occup Med (Oxford, England) 61:78–89. doi:10.1093/occmed/kqr001
Wittmann M, Dinich J, Merrow M, Roenneberg T (2006) Social jetlag: misalignment of biological and social time. Chronobiol Int 23:497–509. doi:10.1080/07420520500545979
Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT (2010) Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol 30:197–205. doi:10.1128/mcb.01154-09
Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH (2005) Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434:640–644. doi:10.1038/nature03453
Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M et al (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152:1091–1105. doi:10.1016/j.cell.2013.01.055
Zelinski EL, Deibel SH, McDonald RJ (2014) The trouble with circadian clock dysfunction: multiple deleterious effects on the brain and body. Neurosci Biobehav Rev 40:80–101. doi:10.1016/j.neubiorev.2014.01.007
Acknowledgments
Our work on the circadian rhythm is funded by the Deutsche Forschungsgemeinschaft through grants HE5398/3 and HE5398/4 to FH.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of a special issue on Micro RNA
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Preußner, M., Heyd, F. Post-transcriptional control of the mammalian circadian clock: implications for health and disease. Pflugers Arch - Eur J Physiol 468, 983–991 (2016). https://doi.org/10.1007/s00424-016-1820-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-016-1820-y