Abstract
Retinoids (natural forms and synthetic derivatives of vitamin A) are used as therapeutic agents for numerous skin diseases such as keratinization disorders (e.g. ichthyoses) and psoriasis. Two endogenous ligands for retinoic acid receptors exist, retinoic acid (atRA) and 3,4-didehydroretinoic acid (ddRA). In primary human epidermal keratinocytes many transcriptional targets for atRA are known, whereas the targets for ddRA are unknown. In an attempt to determine the targets, we compared the effect of atRA and ddRA on transcriptional profiles in undifferentiated and differentiating human primary keratinocytes. First, as expected, many genes were induced or suppressed in response to keratinocyte differentiation. Furthermore, the two retinoids affected substantially more genes in differentiated keratinocytes (>350) than in proliferating keratinocytes (≈20). In differentiating keratinocytes markers of cornification were suppressed suggesting a de-differentiating effect by the two retinoids. When comparing the expression profile of atRA to that of ddRA, no differently regulated genes were found. The array analysis also found that a minor number of miRNAs and a large number of non-coding transcripts were changed during differentiation and in response to the two retinoids. Furthermore, the expression of all, except one, genes known to cause autosomal recessive congenital ichthyosis (ARCI) were found to be induced by differentiation. These results comprehensively document that atRA and ddRA exert similar transcriptional changes in keratinocytes and also add new insights into the molecular mechanism influenced by retinoids in the epidermis. Furthermore, it suggests which ARCI patients could benefit from therapy with retinoids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinoids (natural and synthetic forms of vitamin A) have profound influence on normal keratinocytes both in vitro and in vivo, the best-known effect probably being the inhibition of cellular differentiation. This has resulted in the use of retinoids as therapeutic agents for skin diseases with disturbed keratinization, i.e., ichthyoses and psoriasis [35, 53]. The effects of endogenous vitamin A (retinol) are mediated by its metabolites all-trans retinoic acid (atRA) and 3,4-didehydroretinoic acid (ddRA) [34]. In vitro, retinol is metabolized to atRA and ddRA primarily in differentiated keratinocytes [34, 51]. The keratinocyte-specific ddRA has seemingly similar effects as atRA in some assays [49].
atRA and ddRA exert most of their biological effects by binding to and activating nuclear retinoic acid receptors (RARα, -β and -γ). A second class of retinoid receptors, RXRs (RXRα, -β and -γ), functions as heterodimerization partner for RARs (for review see [30]). These RAR–RXR heterodimers bind to RA-response elements (RAREs) in the promoter region of specific genes (for review see [2]), which lead to altered transcription of the adjacent gene. Several of the retinoid receptors are expressed by human epidermal keratinocytes. The most abundant forms in falling order are RXRα, RARγ and RARα [15], thus making RARγ/RXRα the predominant heterodimer. According to some studies, these receptors are expressed mainly by cells in differentiated cell layers of normal epidermis [25, 36], suggesting that they are involved in the process of terminal differentiation. Yet, another function of unliganded RARs is in the formation of lipid-containing lamellar granules in terminally differentiated keratinocytes [11, 16].
Retinoids also interact with other signaling pathways, e.g., the activator protein-1 (AP-1) (for review see [2]). AP-1 is a regulator of genes induced by differentiation of keratinocytes, e.g., transglutaminase 1 (TGM1), loricrin (LOR), keratin 1 (KRT1), and involucrin (IVL) (see [13] for review), all of which are repressed by atRA [7, 18, 37]. It has also been shown that the DNA-binding activity of AP-1 is increased in cultured keratinocytes induced to differentiate by elevated extracellular calcium [24].
The transcription of many genes is regulated by atRA and a list of 532 genes as regulatory targets of atRA was compiled by evaluating 1,191 published papers [1]. Transcriptional profiling studies in response to atRA have been performed in epidermal keratinocytes and reconstructed skin [4, 26].
The aim of this study was to examine whether the two endogenous retinoids atRA and ddRA affect the mRNA expression profile differently in cultured human primary keratinocytes using oligonucleotide arrays. All genes causing ARCI were analyzed in detail. The expression profiles after retinoid treatment were examined both in proliferating and differentiated keratinocytes, as representatives for basal and suprabasal epidermal keratinocytes, respectively.
Materials and methods
Materials
atRA and ddRA were obtained from Sigma-Aldrich (Stockholm, Sweden) and Roche AG (Basle, Switzerland), respectively. The compounds were dissolved in ethanol and the concentration and purity were determined by UV absorbance and HPLC analysis, respectively. Stock solutions of 1 mM were prepared. The purity of atRA and ddRA was 99.1 and 88.3 %, respectively. The two major impurities in ddRA were 13cRA (1.5 %) and atRA (10.2 %). All handling of retinoids was performed under yellow reduced light.
Cell culture and retinoid treatment
Primary keratinocytes from a lightly pigmented female donor (42 years) were used at passage 3 (Life Technologies, Stockholm, Sweden). The cells were maintained in EpiLife serum-free keratinocyte medium containing growth supplement (HKGS or HKGS kit) and gentamicin/amphotericin (all from Life Technologies). Proliferating cells in 6-well plates at 30–40 % confluency were switched to EpiLife medium without EGF for 5 h prior to starting treatment with retinoids. Keratinocytes at 80 % confluency were switched to EpiLife− supplemented with 1.5 mM CaCl2 for 4 days with the purpose to induce cellular differentiation. Cells were subsequently exposed to retinoids as above in EpiLife medium without rhEGF. Both proliferating and differentiated cells were exposed to 1 µM of atRA, ddRA or vehicle (ethanol) for 4 and 24 h. All treatments were performed in triplicates. The cells were washed with PBS twice and frozen at −70 °C until further processing.
Microarray expression analysis
The cells were dissolved in 1 ml TriReagent (Ambion, Life Technologies, Carlsbad, CA), and total RNA was isolated as previously described [50]. RNA concentration was measured with ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was evaluated using the Agilent 2100 Bioanalyzer system (Agilent Technologies Inc, Palo Alto, CA). Total RNA (250 ng) from each sample was used to generate amplified and biotinylated sense-strand cDNA from the entire expressed genome according to the GeneChip® WT PLUS Reagent Kit User Manual (P/N 703174 Rev 1 Affymetrix Inc., Santa Clara, CA). GeneChip® ST Arrays (GeneChip® XXX Gene 2.1 ST Array) were hybridized for 16 h in a 45 °C incubator, washed and stained and finally scanned at the GeneTitan® Multi-Channel (MC) Instrument, according to the GeneTitan Instrument User Guide for Expression Arrays Plates (PN 702933 Rev 2., Affymetrix Inc., Santa Clara, CA).
Microarray data analysis
The raw data were normalized in the free software Expression Console provided by Affymetrix (http://www.affymetrix.com) using the robust multi-array average (RMA) method [21, 27]. The RMA algorithm fits a robust linear model at the probe level to minimize the effect of probe-specific affinity differences. Subsequent analysis of the gene expression data was carried out in the freely available statistical computing language R (http://www.r-project.org) using packages available from the Bioconductor project (www.bioconductor.org). In order to search for the differentially expressed genes between different groups, an empirical Bayes' moderated t test was then applied [40], using the ‘limma’ package [41]. To address the problem with multiple testing, the p values were adjusted using the method of Benjamini and Hochberg [3].
Heat maps of gene expression were generated using Genesis gene expression similarity investigation suite [44]. Genes were considered regulated if the expression levels differed more than 1.5-fold relative to controls and had an adjusted p value of <0.05. Functional annotation of regulated genes was performed as before using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [19, 20]. Gene ontology clusters with a p value <10−4 were retained for evaluation. The complete data are submitted to Gene Expression Omnibus repository (in process).
Analysis of mRNA expression using quantitative PCR
First-strand cDNA was synthesized from 1.5 µg total RNA by combining oligo(d)T15, random hexamers, buffer and MMLV reverse transcriptase (Life Technologies) as previously described [50].
Semi-quantitative PCR was performed using cDNA (10 ng total RNA) as template and TaqMan Gene Expression Assays and TaqMan Universal PCR Master Mix, No AmpErase UNG in an ABI7500Fast PCR machine (Life Technologies). TaqMan gene expression assays were used to detect TGM1, PNPLA1, LRAT, KRT2, and KRT4. Expression levels were measured in duplicate.
The following genes were analyzed with SYBR Green detection: PPIA-encoding cyclophilin A (used as reference genes for geNorm normalization [54]), TGM1, INL, HB-EGF, ALOX12B, and SCCE. Analysis was performed using an ABI PRISM 7500Fast sequence detection system (Applied Biosystems) as described elsewhere [8, 9]. The relative mRNA expression was determined by the 2(−ΔΔCt) method and analyzed by one-way ANOVA followed by Tukey’s multiple comparison test (Prism 5.04 software, GraphPad Software Inc., La Jolla, CA).
Results
Comparison between differentiating and proliferating primary keratinocytes
We compared the transcriptional profiles of vehicle-exposed differentiating and proliferating keratinocytes using GeneChip® XXX Gene 2.1 ST Arrays. The microarrays simultaneously measure the levels of >41,000 coding and alternative splicing transcript variants and microRNA (miRNA) and long intergenic non-coding transcripts. Of these, approximately ~25,500 coding transcripts, 1,170 miRNA and 17,900 non-coding transcripts were found to be expressed in keratinocytes. Among them were 709 induced and 730 suppressed in differentiating cells, according to our criteria (Table 1). The expressed genes in differentiating and proliferating cells are shown as a heat map (Online Resource 1), and the top ten-induced and -suppressed genes are given in Online Resource 2. Of the induced transcripts were 577 coding and 6 miRNA, and of the suppressed transcripts were 606 coding and 10 miRNA, the remaining transcripts in both groups were non-coding transcripts. Among the many genes affected were 11 genes belonging to the keratin family. KRT1, KRT2, KRT4, KRT6B, KRT6C, KRT10, KRT23, KRT77, and KRT80 were all induced, whereas KRT15 and KRT16 were suppressed. Many of the genes induced by differentiation are also known to be mutated in autosomal recessive congenital ichthyosis (ARCI) or syndromes with congenital ichthyosis, i.e., TGM1, ALOX12B, ALOXE3, CYP4F22, NIPAL4, ABCA12, PNPLA1, CERS3, ABHD5, ALDH3A2, ELOVL4 and SPINK5 (Table 2). One exception was SLC27A4, encoding FATP4.
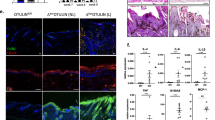
To confirm the oligonucleotide array results obtained, we performed quantitative RT-PCR analysis of genes altered in differentiating vs. proliferating cells or by atRA exposure in differentiating cells. The analysis verified the changes in the mRNA profiling in differentiating vs. proliferating cells (Fig. 1a–c).
Analysis of mRNA expression in proliferating, differentiating and atRA-treated differentiating keratinocytes by quantitative polymerase chain reaction. The mRNA expression of a PNPLA1, b ALOX12B, c TGM1, d KRT2, e KRT4, f INV, g SCCE, h HB-EGF and i LRAT was examined. The expression of PNPLA1, ALOX12B, TGM1 was found to be induced in differentiating vs. proliferating keratinocytes. The expression of TGM1 and KRT2 was significantly reduced in response to atRA treatment in differentiating keratinocytes, whereas the expression of KRT4, INV, SCCE, HB-EGF and LRAT was induced. a–h * p < 0.05, ** p < 0.01, * p < 0.001
Certain miRNAs were also induced (i.e., MIR203, MIR4451, MIR3671, MIR27B, MIR23B) and suppressed (i.e., MIR1305, MIR4435-1, MIR29A, MIR622, MIR181B1, MIRLET7A2, MIR424, MIR503) upon differentiation.
Functional characterization of the genes regulated by differentiation
Functional annotation clustering analysis was performed using DAVID on the genes regulated by differentiation. We found that more than 21 % of the induced genes fall into one of the following ontological categories of biological processes: epidermis development, cell junction, plasma membrane, membrane fraction, cellular lipid metabolic process (Online Resource 3). Among the suppressed genes more than 20 % of them were annotated to the regulation of cell proliferation, blood vessel development, DNA metabolic process, regulation of programmed cell death and regulation of cell cycle.
atRA and ddRA regulate gene expression most profoundly in differentiating keratinocytes
We compared the transcriptional profiles in atRA- and ddRA-treated keratinocytes with vehicle-treated keratinocytes. In cells exposed to one of the two retinoids, the affected RNA transcripts were few in numbers in proliferating keratinocytes as compared to differentiated cells (see Table 1). The genes altered in response to atRA exposure is shown as a heat map (Online Resource 4). However, the number of altered transcripts was in the same range after treatments with both retinoids, and most of the transcripts were significantly altered by both retinoids. The top ten-induced and -suppressed annotated transcripts after atRA exposure are shown in (Online Resource 5). The expression of some genes in atRA- vs. vehicle-treated differentiating cells was verified by qPCR (Fig. 1b–i).
Many of the 151 annotated transcripts induced in differentiating keratinocytes are known to be retinoid regulated, e.g., STRA6, BMP6, LRAT, KRT13, KRT19, KRT23, KRT31, KRT4, DHRS3, DHRS9, KLK6, HB-EGF, S100A7 (PSORIASIN) and PI3 (SKALP). Among the ones not previously reported were GBP6, GPR176, SERPINB1, SERPINB3, STAT4, and CLEC2. Some of the 143 annotated transcripts suppressed in differentiating keratinocytes are known to be affected by retinoids, e.g., KRT1, KRT2, FLG and TGM1. The effect on the expression of ARCI-causing genes is given in Table 2. Among the ones not previously reported were GPR155, CDHR1, FLG2, and ACER1. There was also altered expression of a few miRNAs. MIR1305 and MIR3975 were induced and MIR203, MIR4774, MIR3671, and MIR4451 were suppressed by atRA exposure.
Functional characterization of the atRA-regulated genes in differentiating keratinocytes
When we summarized the functional categories of the genes up-regulated by atRA at 24 h in differentiated keratinocytes, we found that 11 % of them fall into the categories tissue development or hormone metabolic process (Online Resource 6). Among the suppressed genes, more than 10 % of them were annotated to either lipid catabolic process or vesicular fraction.
When combining all up-regulated and suppressed genes about 40 % of the genes were related to plasma membrane, endoplasmic reticulum, cell–cell junction, endopeptidase activity and intermediate filament (Online Resource 6).
Gene expression was not regulated differently by atRA and ddRA
Although the major number of retinoid-regulated transcripts was induced or suppressed equally efficiently by atRA and ddRA, there were also transcripts that were significantly altered by only one of the retinoids when comparing to vehicle-treated controls. However, when comparing the expression profiles generated by the two retinoids with each other, no genes were significantly altered (data not shown).
Discussion
In the present study, we examined gene expression profiles in cultured keratinocytes. First, we examined the expression profiles in proliferating and differentiating keratinocytes and secondly, the effects of exogenous addition of two retinoids which normally are synthesized by epidermal keratinocytes, atRA and ddRA. The reason for comparing the expression profile in both cell types is that retinoid receptors are to a major extent expressed by keratinocytes in suprabasal layers of epidermis [25, 36], suggesting that retinoids may affect gene transcription mainly in differentiated cells.
We found that the genes altered by differentiation were involved in epidermis development, plasma membrane, cellular lipid metabolic process, regulation of cell proliferation, blood vessel development, DNA metabolic process and regulation of DNA metabolic process. Our results verify the results by others regarding many of the altered genes [26]. Several of the genes are regulated during keratinocyte differentiation [26, 45].
Using a macro-array methodology, it has been shown that retinoids generate different effects when added to reconstructed human epidermis as compared to cultured keratinocytes [4]. In these experiments, the cultured cells were not induced to differentiate and thus the cells are equivalent to the proliferating cells in the present experiments.
Others have found that atRA suppresses markers of cornification and genes involved in de novo lipogenesis [26]. Also, at RA was shown to regulate the pathways of its own bioavailability and genes associated with the cell cycle and programmed cell death [26]. In the present study, we found regulation of pathways related to cornification genes, few genes involved in atRA bioavailability and programmed cell death but not of genes involved in de novo lipogenesis. The lack of effect on the expression of these genes could be due to difference between the experiments with respect to the number of time points investigated and the culture conditions (proliferating vs. differentiating) [26]. In the present study, differentiation was achieved by increasing calcium concentration for the last 3 days and removing EGF for the last 5 h prior to addition of retinoids to the medium.
Autosomal recessive congenital ichthyosis is to date known to be caused by ≥10 genes [46]. Many of these genes are involved in transport or metabolism of epidermal lipids, whereas other genes are involved in modifying omega-hydroxy-glucosylceramides for covalent binding to the cornified lipid envelope, a step which is performed by transglutaminase-1 (encoded by TGM1) [14]. We found several of the genes causing ARCI and other forms of ichthyosis to be induced during keratinocyte differentiation, i.e., TGM1, ALOXE3, ALOX12B, NIPAL4, CYP4F22, ABCA12, ABHD5, PNPLA1, ELOVL4, LIPN and CERS3 (see Table 2). The present results correlate well with the previously reported expression in cultured keratinocytes [12, 23, 31, 42] and localization of respective protein in upper layers of normal epidermis [10, 17, 28, 29, 33, 38, 39]. However, to the best of our knowledge, this is the first time the expression of ALOX12B, ALOXE3, LIPN, CYP4F22, PNPLA1 and ELOVL4 has been studied in cultured human keratinocytes. Surprisingly, SLC27A4 transcripts were not induced by differentiation. Furthermore, some of the transcripts were significantly suppressed by retinoids, i.e., ALOX12B, PNPLA1, and LIPN (see Table 2).
It was recently shown that keratinocyte differentiation is positively regulated by the microRNA mir-203 [43]. MIR203 (and MIR3671) were induced by differentiation in our array analysis supporting the robustness of the present data, with respect to miRNAs. Furthermore, both MIR203 and MIR3671 expressions were suppressed by atRA exposure in differentiated keratinocytes. This suggests that also mir-3671 might be a positive regulator of keratinocyte differentiation. On the other hand, mir-1305 expression was altered in an opposite manner compared to these two miRNAs.
The role of 3,4-didehydroretinoids (3,4-didehydroretinol, 3,4-didehydroretinaldehyde and 3,4-didehydroretinoic acid) is unclear. In mouse keratinocytes, it has been shown that apoptosis is induced to greater extent by ddRA compared to atRA [22]. This effect on apoptosis is supported by induction of genes annotated to apoptosis after exposure to atRA as previously described [26]. However, in the present study no difference in transcription profiles was found between atRA and ddRA at any time point.
It is possible that these two endogenous retinoids do not have different functions or that one should look at different aspects of biological function, not directly related to gene transcription. In other organisms, 3,4-didehydroretinaldehyde, alone or in combination with retinaldehyde, is used in the retinal chromophore [32, 47, 48, 52]. In Gecko lizards it has been shown that in the eye lens CRBPI has 3,4-didehydroretinol as ligand which gives the lens a yellow color [55]. It has been suggested that 3,4-didehydroretinoids in the retina diminishes the harmful effects of short-wave radiation [55]. Furthermore, in human and rabbit skin it has been shown that exposure to UVB light degrades retinol, but not 3,4-didehydroretinol [5, 6]. This may suggest that the biosynthesis of epidermal 3,4-didehydroretinoids serves as a back-up system for retinol and retinoic acid during situations of exposure to UV radiation.
In summary, these data present a comprehensive description of the transcriptional changes caused by atRA and ddRA treatment of proliferating and differentiating primary human keratinocytes. The transcriptional changes obtained by the two retinoids were exactly the same, ruling out different transcriptional functions for these two endogenous retinoids formed by epidermal keratinocytes. We also found that the expression of 13 genes causing ARCI and syndromes with ichthyosis is induced in differentiating keratinocytes, and four of these genes showed reduced expression upon atRA treatment. The effect of retinoids in the treatment of ARCI patients could thus be due to inhibition of the expression of the disease-causing genes or a general effect on keratinocyte differentiation. Surprisingly, far from all ARCI-causing genes were suppressed by atRA exposure arguing against a de-differentiation effect. Hypothetically, responders to retinoid therapy might be found among those patients with mutations in genes that are not affected by retinoids in vitro (due to suppression of residual enzymatic activity).
References
Balmer JE, Blomhoff R (2002) Gene expression regulation by retinoic acid. J Lipid Res 43(11):1773–1808
Bastien J, Rochette-Egly C (2004) Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1–16
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57(1):289–300
Bernard FX, Pedretti N, Rosdy M, Deguercy A (2002) Comparison of gene expression profiles in human keratinocyte mono-layer cultures, reconstituted epidermis and normal human skin; transcriptional effects of retinoid treatments in reconstituted human epidermis. Exp Dermatol 11(1):59–74
Berne B, Nilsson M, Vahlquist A (1984) UV irradiation and cutaneous vitamin A: an experimental study in rabbit and human skin. J Invest Dermatol 83:401–404
Berne C, Vahlquist A, Fischer T, Danielsson B, Berne B (1984) UV treatment of uraemic pruritus reduces the vitamin A content of the skin. Eur J Clin Invest 14:203–206
Brown LJ, Geesin JC, Rothnagel JA, Roop DR, Gordon JS (1994) Retinoic acid suppression of loricrin expression in reconstituted human skin cultured at the liquid-air interface. J Invest Dermatol 102(6):886–890
Buraczewska I, Berne B, Lindberg M, Loden M, Törmä H (2009) Moisturizers change the mRNA expression of enzymes synthesizing skin barrier lipids. Arch Dermatol Res 301:587–594
Buraczewska I, Berne B, Lindberg M, Lodén M, Törmä H (2009) Long-term treatment with moisturizers affects the mRNA levels of genes involved in keratinocyte differentiation and desquamation. Arch Dermatol Res 301:175–181
Buxman MM, Wuepper KD (1978) Cellular localization of epidermal transglutaminase: a histochemical and immunochemical study. J Histochem Cytochem 26(5):340–348
Calleja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, Metzger D, Mascrez B, Ohta K, Kagechika H, Endo Y, Mark M, Ghyselinck NB, Chambon P (2006) Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev 20(11):1525–1538. doi:10.1101/gad.368706
Dahlqvist J, Westermark GT, Vahlquist A, Dahl N (2012) Ichthyin/NIPAL4 localizes to keratins and desmosomes in epidermis and Ichthyin mutations affect epidermal lipid metabolism. Arch Dermatol Res 304(5):377–386. doi:10.1007/s00403-012-1207-7
Eckert RL, Welter JF (1996) Transcription factor regulation of epidermal keratinocyte gene expression. Mol Biol Rep 23(1):59–70
Elias PM, Gruber R, Crumrine D, Menon G, Williams ML, Wakefield JS, Holleran WM, Uchida Y (2014) Formation and functions of the corneocyte lipid envelope (CLE). Biochim Biophys Acta 1841(3):314–318. doi:10.1016/j.bbalip.2013.09.011
Fisher GJ, Talwar HS, Xiao JH, Datta SC, Reddy AP, Gaub MP, Rochette-Egly C, Chambon P, Voorhees JJ (1994) Immunological identification and functional quantitation of retinoic acid and retinoid X receptor proteins in human skin. J Biol Chem 269(32):20629–20635
Ghyselinck N, Chambon P (2007) Animal models for retinoid receptor research; implications for epidermal homeostasis, skin barrier function, wound healing and atopic dermatitis. In: Vahlquist A, Duvic M (eds) Retinoids and carotenoids in dermatology. Informa Healthcare USA Inc., New York, pp 27–54
Grall A, Guaguere E, Planchais S, Grond S, Bourrat E, Hausser I, Hitte C, Le Gallo M, Derbois C, Kim GJ, Lagoutte L, Degorce-Rubiales F, Radner FP, Thomas A, Kury S, Bensignor E, Fontaine J, Pin D, Zimmermann R, Zechner R, Lathrop M, Galibert F, Andre C, Fischer J (2012) PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet 44(2):140–147. doi:10.1038/ng.1056
Griffiths CEM, Rosenthal DS, Reddy AP, Elder JT, Astrom A, Leach K, Wang TS, Finkel LJ, Yuspa SH, Voorhees JJ, Fisher GJ (1992) Short-term retinoic acid treatment increases in vivo, but decreases in vitro, epidermal transglutaminase-K enzyme activity and immunoreactivity. J Invest Dermatol 99:283–288
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. doi:10.1093/nar/gkn923
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. doi:10.1038/nprot.2008.211
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4(2):249–264. doi:10.1093/biostatistics/4.2.249
Islam TC, Skarin T, Sumitran S, Toftgard R (2000) Retinoids induce apoptosis in cultured keratinocytes. Br J Dermatol 143(4):709–719
Jiang YJ, Lu B, Kim P, Paragh G, Schmitz G, Elias PM, Feingold KR (2008) PPAR and LXR activators regulate ABCA12 expression in human keratinocytes. J Invest Dermatol 128(1):104–109
Johansen C, Iversen L, Ryborg A, Kragballe K (2000) 1α,25-Dihydroxyvitamin D3 induced differentiation of cultured human keratinocytes is accompanied by a PKC-independent regulation of AP-1 DNA binding activity. J Invest Dermatol 114(6):1174–1179
Karlsson T, Rollman O, Vahlquist A, Törmä H (2004) Immunofluorescence localization of nuclear retinoid receptors in psoriasis versus normal human skin. Acta Derm Venereol 84(5):363–369
Lee DD, Stojadinovic O, Krzyzanowska A, Vouthounis C, Blumenberg M, Tomic-Canic M (2009) Retinoid-responsive transcriptional changes in epidermal keratinocytes. J Cell Physiol 220(2):427–439. doi:10.1002/jcp.21784
Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98(1):31–36. doi:10.1073/pnas.011404098
Li H, Pavez Loriè E, Fisher J, Vahlquist A, Törmä H (2012) The expression of epidermal lipoxygenases and transglutaminase-1 is perturbed by NIPAL4 mutations: indications of a common metabolic pathway essential for skin barrier homeostasis. J Invest Dermatol 132:2368–2375. doi:10.1038/jid.2012.160
Li H, Vahlquist A, Törmä H (2013) Cross-talk between FATP4 and ichthyin in epidermal lipid processing? Clues from studies of two types of autosomal recessive congenital ichthyosis. J Dermatol Sci 69:195–201. doi:10.1016/j.jdermsci.2012.11.593
Mark M, Ghyselinck NB, Chambon P (2006) Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol 46:451–480. doi:10.1146/annurev.pharmtox.46.120604.141156
Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y (2008) 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J Lipid Res 49(11):2356–2364. doi:10.1194/jlr.M800158-JLR200
Okano K, Oishi T, Miyashita Y, Moriya T, Tsuda M, Irie T, Ueki N, Seki T (2002) Identification of 3,4-didehydro retinal isomers in the Xenopus tadpole tail fin containing photosensitive melanophores. Zoolog Sci 19(2):191–195
Radner FP, Marrakchi S, Kirchmeier P, Kim GJ, Ribierre F, Kamoun B, Abid L, Leipoldt M, Turki H, Schempp W, Heilig R, Lathrop M, Fischer J (2013) Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet 9(6):e1003536. doi:10.1371/journal.pgen.1003536
Randolph RK, Simon M (1993) Characterization of retinol metabolism in cultured human epidermal keratinocytes. J Biol Chem 268(13):9198–9205
Raut AS, Prabhu RH, Patravale VB (2013) Psoriasis clinical implications and treatment: a review. Crit Rev Ther Drug Carrier Syst 30(3):183–216
Reichrath J, Mittmann M, Kamradt J, Muller SM (1997) Expression of retinoid-X receptors (-alpha,-beta,-gamma) and retinoic acid receptors (-alpha,-beta,-gamma) in normal human skin: an immunohistological evaluation. Histochem J 29(2):127–133
Rosenthal DS, Griffiths CE, Yuspa SH, Roop DR, Voorhees JJ (1992) Acute or chronic topical retinoic acid treatment of human skin in vivo alters the expression of epidermal transglutaminase, loricrin, involucrin, filaggrin, and keratins 6 and 13 but not keratins 1, 10, and 14. J Invest Dermatol 98(3):343–350
Sakai K, Akiyama M, Sugiyama-Nakagiri Y, McMillan JR, Sawamura D, Shimizu H (2007) Localization of ABCA12 from Golgi apparatus to lamellar granules in human upper epidermal keratinocytes. Exp Dermatol 16(11):920–926. doi:10.1111/j.1600-0625.2007.00614.x
Sasaki K, Akiyama M, Yanagi T, Sakai K, Miyamura Y, Sato M, Shimizu H (2012) CYP4F22 is highly expressed at the site and timing of onset of keratinization during skin development. J Dermatol Sci 65(2):156–158. doi:10.1016/j.jdermsci.2011.12.006
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. doi:10.2202/1544-6115.1027
Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, pp 397–420
Song HJ, Rossi A, Ceci R, Kim IG, Anzano MA, Jang SI, De Laurenzi V, Steinert PM (1997) The genes encoding geranylgeranyl transferase alpha-subunit and transglutaminase 1 are very closely linked but not functionally related in terminally differentiating keratinocytes. Biochem Biophys Res Commun 235(1):10–14
Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Törmä H, Ståhle M, Pivarcsi A (2010) Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol 130(1):124–134
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18(1):207–208
Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, de Daruvar A, Wincker P, Serre G, Guerrin M (2007) Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol 8(6):R107
Traupe H, Fischer J, Oji V (2014) Nonsyndromic types of ichthyoses––an update. J Dtsch Dermatol Ges 12(2):109–121. doi:10.1111/ddg.12229
Tsin AT, Alvarez RA, Fong SL, Bridges CD (1985) Conversion of retinol to 3,4-didehydroretinol in the tadpole. Comp Biochem Physiol B 81(2):415–421
Tsin AT, Santos FR (1985) The 3, 4-didehydroretinal chromophore of goldfish porphyropsin. J Exp Zool 235(2):181–186. doi:10.1002/jez.1402350204
Törmä H, Asselineau D, Andersson E, Martin B, Reiniche P, Chambon P, Shroot B, Darmon M, Vahlquist A (1994) Biologic activities of retinoic acid and 3,4-didehydroretinoic acid in human keratinocytes are similar and correlate with receptor affinities and transactivation properties. J Invest Dermatol 102(1):49–54
Törmä H, Geijer S, Gester T, Alpholm K, Berne B, Lindberg M (2006) Variations in the mRNA expression of inflammatory mediators, markers of differentiation and lipid-metabolizing enzymes caused by sodium lauryl sulphate in cultured human keratinocytes. Toxicol In Vitro 20(4):472–479
Törmä H, Rollman O, Vahlquist A (1999) The vitamin A metabolism and expression of retinoid-binding proteins differ in HaCaT cells and normal human keratinocytes. Arch Dermatol Res 291(6):339–345
Ueno Y, Ohba H, Yamazaki Y, Tokunaga F, Narita K, Hariyama T (2005) Seasonal variation of chromophore composition in the eye of the Japanese dace, Tribolodon hakonensis. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 191(12):1137–1142. doi:10.1007/s00359-005-0037-x
Vahlquist A, Gånemo A, Virtanen M (2008) Congenital ichthyosis: an overview of current and emerging therapies. Acta Derm Venereol 88(1):4–14
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034
Werten PJ, Roll B, van Aalten DM, de Jong WW (2000) Gecko iota-crystallin: how cellular retinol-binding protein became an eye lens ultraviolet filter. Proc Natl Acad Sci USA 97(7):3282–3287
Acknowledgements
This study was performed with support from The Welander and Finsen foundation and Swedish Research Council (project: K2013-57X-22309-01-3). We are grateful for the technical assistance with DNA microarrays and data analysis provided by the Array and Analysis Facility, Science for Life Laboratory at Uppsala Biomedical Centre (BMC), Husargatan 3, 751 23 Uppsala.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Törmä, H., Bergström, A., Ghiasifarahani, G. et al. The effect of two endogenous retinoids on the mRNA expression profile in human primary keratinocytes, focusing on genes causing autosomal recessive congenital ichthyosis. Arch Dermatol Res 306, 739–747 (2014). https://doi.org/10.1007/s00403-014-1476-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00403-014-1476-4