Abstract
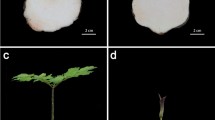
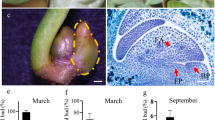
Aechmea fasciata is a well-known ornamental flowering plant in the bromeliad family. It is proposed that a small burst of ethylene synthesis in the meristem triggers flowering in pineapple and other bromeliads in response to diverse environmental and endogenous signals. A. fasciata showed an age-dependent response: adult plants were induced to flower successfully under ethylene treatment but juvenile plants did not. To better understand the mechanism of different responses to ethylene in transcriptome and flowering induction by ethylene, we performed a comparative analysis of A. fasciata transcriptome. Four libraries of A. fasciata adult and juvenile plants under water and ethylene treatment were sequenced by Illumina deep sequencing, 55,238,936, 53,797,292, 53,471,812, and 53,485,862 qualified Illumina clean reads, respectively, with 90 bp mean length, respectively. Unigenes of the four libraries were assembled and then merged into an unified library with 86,609 sequences and a mean size of 987 bp. After searching against Nr, KEGG, Swiss-Prot, and COG databases, 28,350 sequences were assigned to 129 KEGG pathways, 28,289 unigenes were categorized into 64 functional groups, and 19,293 sequences were classified into 25 COG categories. Through differential expression analysis, 56 DEGs related to flowering were identified. The critical genes correlated with flowering were selected and confirmed by qRT-PCR analysis. This study provided a global survey of changes in transcriptomes of A. fasciata in response to ethylene. The analyses of transcriptome profiles imply that FT is upregulated in the adult plant and results in flowering. Moreover, the differential expression of GI, DELLA, GAD1, AP2, and so on indicated that a complicated network participated in the induction of flowering by ethylene, which will help in the future studies.








Similar content being viewed by others
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y et al (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P et al (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104:6484–6489
Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z et al (2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant cell 13:2455–2470
Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB et al (2007) The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19:509–523
Bolouri Moghaddam MR, den Ende WV (2013) Sugars, the clock and transition to flowering. Fron Plant Sci 4:22. doi:10.3389/fpls.2013.00022
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM et al (2004) GO:TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715
Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D et al (2007) A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol 145:98–105
Chuck G, Hake S (2005) Regulation of developmental transitions. Curr Opin Plant Biol 8:67–70
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Davis SJ (2009) Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ 32:1201–1210
de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S et al (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451:480–484
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141:550 e551–550 e552
Galvao VC, Horrer D, Kuttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139:4072–4082
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94:261–271
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting evaluating and reconstructing potential coding regions in EST sequences. ISMB 99:138–148
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH et al (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19:2736–2748
Jung CH, Wong CE, Singh MB, Bhalla PL (2012) Comparative genomic analysis of soybean flowering genes. PLoS One 7:e38250
Jung JH, Lee S, Yun J, Lee M, Park CM (2014) The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci 215–216:29–38
Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72:427–441
Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP et al (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484:242–245
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E et al (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23:1815–1829
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods 25:402–408
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793
Matsoukas IG (2014) Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Frontiers in genetics 5:218
Merchante C, Alonso JM, Stepanova AN (2013) Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 16:554–560
Middleton AM, Ubeda-Tomas S, Griffiths J, Holman T, Hedden P, Thomas SG et al (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci USA 109:7571–7576
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Mutasa-Gottgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60:1979–1989
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45
Pineiro M, Jarillo JA (2013) Ubiquitination in the control of photoperiodic flowering. Plant science : an international journal of experimental plant biology 198:98–109
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108:11698–11703
Schaller GE (2012) Ethylene and the regulation of plant development. BMC Biol 10:9
Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12:3703–3714
Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18:575–583
Trusov Y, Botella JR (2006) Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.]. J Exp Bot 57:3953–3960
Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A et al (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707
Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64:675–684
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A et al (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU et al (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Yu S, Galvao VC, Zhang YC, Horrer D, Zhang TQ, Hao YH et al (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24:3320–3332
Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q et al (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106:21431–21436
Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21:841–847
Acknowledgments
The research was supported by the The National Natural Science Foundation of China (31372106) and The Fundamental Scientific Research Funds for CATAS-TCGRI (1630032014018).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhiying Li and Jiabin Wang have been contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Wang, J., Zhang, X. et al. Transcriptome Sequencing Determined Flowering Pathway Genes in Aechmea fasciata Treated with Ethylene. J Plant Growth Regul 35, 316–329 (2016). https://doi.org/10.1007/s00344-015-9535-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-015-9535-4