Abstract
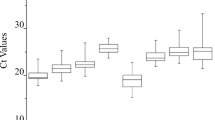
To screen the stable expression genes related to the stress (strong light, dehydration and temperature shock) we applied Absolute real-time PCR technology to determine the transcription numbers of the selected test genes in P orphyra yezoensis, which has been regarded as a potential model species responding the stress conditions in the intertidal. Absolute real-time PCR technology was applied to determine the transcription numbers of the selected test genes in P orphyra yezoensis, which has been regarded as a potential model species in stress responding. According to the results of photosynthesis parameters, we observed that Y(II) and F v/F m were significantly affected when stress was imposed on the thalli of P orphyra yezoensis, but underwent almost completely recovered under normal conditions, which were collected for the following experiments. Then three samples, which were treated with different grade stresses combined with salinity, irradiation and temperature, were collected. The transcription numbers of seven constitutive expression genes in above samples were determined after RNA extraction and cDNA synthesis. Finally, a general insight into the selection of internal control genes during stress response was obtained. We found that there were no obvious effects in terms of salinity stress (at salinity 90) on transcription of most genes used in the study. The 18S ribosomal RNA gene had the highest expression level, varying remarkably among different tested groups. RPS8 expression showed a high irregular variance between samples. GAPDH presented comparatively stable expression and could thus be selected as the internal control. EF-1α showed stable expression during the series of multiple-stress tests. Our research provided available references for the selection of internal control genes for transcripts determination of P. yezoensis.
Similar content being viewed by others
Abbreviations
- 18S :
-
18S rRNA
- RPS8 :
-
30S ribosomal protein S8
- PUB-2 :
-
polyubiquitin-2
- GAPDH :
-
glyceraldehyde-3-phosphate dehydrogenase
- EF1-α :
-
elongation factor1-α
- TubB :
-
betatubulin
- Act2 :
-
actin2
- PAM:
-
pulse amplitude modulated fluorometry
References
Andersen C L, Jensen J L, Ørntoft T F. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Re search, 64: 5 245–5 250.
Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B. 1989. The gene family encoding the Arabidopsis thaliana translation elongation factor EF-lα: molecular cloning, characterization and expression. Molecular Genetics and Genomics, 219: 106–112.
Beer S, Larsson C, Poryan O, Axelsson L. 2000. Photosynthetic rates of Ulva (Chlorophyta) measured by pulse amplitude modulated (PAM) fluorometry. European Journal of Phycology, 35: 69–74.
Binet M, Weil J, Tessier L. 1991. Structure and expression of sunflower ubiquitin genes. Plant Molecular Biology, 17: 395–407.
Blouin N A, Brodie J A, Grossman A C, Xu P, Brawley S H. 2010. Porphyra: a marine crop shaped by stress. Trends in Plant Science, 16: 29–37.
Bustin S A. 2000. Absolute quantification of mRNA using realtime reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology, 25: 169–193.
Bustin S A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of Molecular Endocrinology, 29: 23–39.
Christensen A H, Quail P H. 1989. Sequence analysis and transcriptional regulation by heat shock of polyubiquitin transcripts from maize. Plant Molecular Biology, 12: 619–632.
Dharmawardhane S, Demma M, Yang F, Condeelis J. 1991. Compartmentalization and actin binding-properties of ABP-50-the elongation factor-1alpha of Dictyostelium. Cell Motil Cytoskeleton, 20: 279–288.
Gao S, Wang G. 2012. The enhancement of cyclic electron flow around photosystem I improves the recovery of severely desiccated Porphyra yezoensis (Bangiales, Rhodophyta). Journal of E xperimental B otany, 63: 4 349–4 358.
Genty B, Briantais J M, Baker N R. 1989. The relationship between the quantum yield of photosynthetic electrontransport and quenching of chlorophyll fluorescence. Biochimica et Biophysica Acta, 990: 87–92.
Jain M A, Nijhawan A, Tyagi A K, Khurana J P. 2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications, 345: 646–651.
Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert H J. 2001. Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell, 13: 889–905.
Marri L, Sparla F, Pupillo P, Trost P. 2005. Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. Journal of Experimental Botany, 56: 73–80.
Öquist G, Fork D C. 1982. Effects of desiccation on the excitation energy distribution from phycoerythrin to the two photosystems in the red alga Porphyra perforata. Physiologia Plantarum, 56: 56–62.
Ostrem J A, Vernon D M, Bohnert H J. 1990. Increased expression of a gene coding for NAD: glyceraldehyde-3-phosphate dehydrogenase during the transition from C3 photosynthesis to crassulacean acid metabolism in Mesembryanthemum crystallinum. Journal of Biochemistry and Molecular Biology, 265: 3 497–3 502.
Prasil O, Suggett D J, Cullen J J, Govindjee B M. 2008. Aquafluo 2007: chlorophyll fluorescence in aquatic sciences, an international conference held in Nové Hrady. Photosynthesis Research, 95: 111–115.
Russell D A, Sachs M M. 1989. Differential expression and sequence analysis of the maize glyceraldehyde-3-phosphate dehydrogenase gene family. The Plant Cell, 1: 793–803.
Schreiber U, Schliwa U, Bilger W. 1986. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynthesis Research, 10: 51–62.
Setter T L, Flannigan B A. 2001. Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. Journal of Experimental Botany, 52: 1 401–1 408.
Suzuki T, Higgins P J, Crawford D R. 2000. Control selection for RNA quantitation. Biotechniques, 29: 332–337.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, Paepe A D, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3: RESEARCH0034.
Whelan J A, Russell N B, Whelan M A. 2003. A method for the absolute quantification of cDNA using real-time PCR. Journal of Immunological Methods, 278: 261–269.
Wong M L, Medrano J F. 2005. Real-time PCR for mRNA quantitation. BioTechniques, 39: 75–85.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supported by the National Natural Science Foundation of China (Nos. 41176134, 30830015), the Prospective Joint Research Project of Jiangsu Province (No. BY2011188), the National Marine Public Welfare Research Project (Nos. 201105008-2, 201105023-7), and the National Basic Research Program of China (973 Program) (No. 2011CB411908)
Rights and permissions
About this article
Cite this article
Wang, W., Wu, X., Wang, C. et al. Exploring valid internal-control genes in Porphyra yezoensis (Bangiaceae) during stress response conditions. Chin. J. Ocean. Limnol. 32, 783–791 (2014). https://doi.org/10.1007/s00343-014-3245-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-014-3245-9