Abstract
Non-indigenous species (NIS) in the Arctic have an increased likelihood of arrival from ship traffic in the region, while the survival potential of the species becomes more likely in a warming environment. Monitoring is essential to detect the rate and magnitude of the establishment of NIS. In this study, a list of 123 potential marine NIS for Svalbard was drafted and the presence of marine NIS in soft sediment of Kongsfjorden in Svalbard was assessed using molecular metabarcoding techniques. For 37 species, including eight potential Arctic NIS, we generated new 18S and/or COI barcode sequences to improve the available online reference databases. In total, 299 species were identified in the sediment samples, including seven potential NIS. Three of these potential NIS have not been reported before in Svalbard: the harpacticoid copepod Euterpina acutifrons, and the ascidians Botrylloides violaceus and Molgula manhattensis. Another novel observation for Svalbard was the polychaete Chone mollis. Additional studies are needed to assess whether the NIS have been established on Svalbard and what their potential impact on the local system may be. Metabarcoding proved to be an effective monitoring tool to detect the presence of new species in Svalbard marine waters. We advise its use to set up a baseline record for the presence of NIS at points of entry, especially harbours. This approach is also valuable for biodiversity monitoring, in particular the detection of small organisms and life stages that are hard to identify using current visual techniques.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-indigenous species (NIS) have the potential to transform local ecosystems by changing the community structure, altering fundamental ecosystem processes or changing the physical environment, with potentially large environmental and economic consequences (Bax et al. 2003; Molnar et al. 2008). Early detection is essential for NIS management (Simberloff et al. 2005; Trebitz et al. 2017), especially when vectors of introduction can be rapidly identified and mitigated.
With the Arctic experiencing its highest rate of global warming over the past decades, NIS are more able than ever to arrive and establish in regions in which they previously could not survive and reproduce (Walther et al. 2009). The risk of introducing marine NIS is particularly high in environments where shipping traffic and the discharge of ballast water present efficient vectors for their spread (Brown et al. 2016). A closer monitoring of possible vectors for the introduction of NIS in the Arctic is recommended to be prepared for development of potential pests (Goldsmit et al. 2014). Chan et al. (2018) reported 54 introductions involving 34 NIS in the Arctic region, 26 of which have led to established populations in Arctic waters. As the Arctic includes a variety of habitats and covers an extensive area, species endemic to one part of the Arctic may be non-indigenous to others. The introduction of NIS in the Arctic, therefore, can result not only from transport to the Arctic, but also from within the Arctic region.
A comprehensive baseline of the current biodiversity in an area is crucial to enable the assessment of changes taking place in the biodiversity as well as the identification of potential anthropogenic routes of introduction of NIS. Both are needed for the development of a strategy for the protection of native species from human-facilitated invasions of NIS (Goldsmit et al. 2014). Research on NIS is ongoing in several areas of the Arctic (CAFF 2013; Reimer et al. 2017). The Circumpolar Biodiversity Monitoring Program (CBMP) of the Arctic Council working group on Conservation of Arctic Flora and Fauna (CAFF) has developed a program to establish a biodiversity baseline and monitor change in species composition of coastal and open-sea habitats of the Arctic. The efforts of the CBMP, which include ongoing scientific projects and studies of time series across the Arctic, are an important step toward resolving which species are native, thereby permitting the identification of new species.
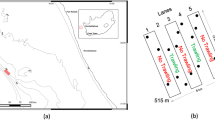
Svalbard is an island archipelago located north of Norway (Fig. 1). Recent warming of the waters west and north of Svalbard is resulting in more suitable habitat conditions for the settlement of non-native species (Førland et al. 2011; Węsławski et al. 2018; Leopold et al. 2019). New marine species may arrive in Svalbard either by natural dispersion, by human assistance (hull fouling, ballast water) or a combination of both. Mussels of the genus Mytilus reappeared on Svalbard between 2000 and 2004 after 1000 years of absence (Berge et al. 2005; Leopold et al. 2019). This was suggested to be either due to a fluctuating slow natural range extension (Węsławski et al. 2011), a single exceptional warm inflow of Atlantic water carrying larvae north (Berge et al. 2005), and/or adult specimens transported to Svalbard by rafted debris or ship traffic (Węsławski and Kotwicki 2018; Leopold et al. 2019). Other examples of temperate and boreal species being transported northwards with the extension of the North Atlantic Current are the euphausiid Meganyctiphanes norvegica (Buchholz et al. 2010) and the starfish Asterias rubens (Deja et al. 2016). The Norwegian Biodiversity Information Centre has identified 31 species to be ‘doorstep species’ for Svalbard. These are species not yet established in the area but expected to settle within the next 50 years through various vectors of introduction (https://www.artsdatabanken.no). Of the 24 marine species on this list, 23 were identified as likely to soon become established in Svalbard. The list covered NIS that are likely to expand to Svalbard based on their current distribution and did not include NIS that need transport over long distances.
Information on the actual presence of marine NIS in the Svalbard environment is currently sparse and anecdotal. In the bordering Barents Sea, seven NIS were reported as established in the area: the arthropods Caprella mutica, Eurytemora americana, Chionoecetes opilio, Eriocheir sinensis and Paralithodes camtschaticus, the bivalve Mytilus galloprovincialis, and the platyhelminth Gyrodactylus salaris (Chan et al. 2018). The Alaskan king crab (P. camtschaticus) has an established population in the Barents Sea. It was introduced deliberately for fisheries purposes in the 1960s (Jørgensen et al. 2005; Ruiz and Hewitt 2009). The snow crab (C. opilio) was first observed in Svalbard waters in 2017 and was probably unintentionally introduced into the Barents Sea via ballast water before spreading throughout the region (Berge et al. 2017; Chan et al. 2018). Pacific pink salmon (Oncorhynchus gorbuscha) escaped from Murmansk cage cultures and was found on Svalbard in the 1960s; it may now be spawning in the region (Witkowski and Glowacki 2010). Some studies have reported the presence of marine NIS on Svalbard as biofouling on ship hulls (e.g. cargo and tourist vessels), ballast water (of coal bulk carriers), and on floating debris (Ware et al. 2016; Węsławski and Kotwicki 2018). Multiple non-indigenous zooplankton species (23) were identified from ship ballast water discharged near Svalbard ports (Ware et al. 2016), though none of these have as of yet been reported as being established in the Svalbard environment.
The main reason for the paucity of information on NIS in Svalbard marine waters is a lack of effective and rapid techniques for monitoring marine biodiversity along the extensive coast. Traditional, morphological identification methods are labour intensive and require specialist knowledge, and species detection is low when populations occur at low densities. Moreover, visual identification techniques often do not allow identification of juvenile life stages of benthic or pelagic animals because species characteristics have not yet been fully developed. Identifying these juvenile stages, however, is critically important for monitoring the establishment and spread of NIS populations (Brown et al. 2016). Many sessile marine species can only disperse as juvenile stages in the water column before settlement and therefore signify successful recruitment.
Molecular technology can offer an efficient approach to the identification of NIS (Chan et al. 2018). In particular, metabarcoding of DNA extracted from environmental samples (environmental DNA or eDNA) enables the simultaneous detection of multiple species (Jeunen et al. 2019; Van der Loos and Nijland 2020). This method allows the identification of DNA contained in live-collected organisms, but also in fragments that animals release into their environment (Jeunen et al. 2019). It is useful in monitoring the presence of species in water, soil or air samples (Leray et al. 2013), and especially in cold water regions where DNA degradation is slower due to low temperatures (Lacoursière-Roussel et al. 2018). DNA metabarcoding may therefore provide a valuable alternative method for the identification of NIS living in the direct vicinity of the sampled site compared with traditional methods (Holman et al. 2019). Yet, despite the increasing number of studies using eDNA metabarcoding for the marine realm, the field is still in development and optimisation of protocols for e.g. sampling, DNA conservation and extraction, barcode selection as well as data analysis is ongoing (Ruppert et al. 2019). Important challenges in eDNA metabarcoding include the spatial relevance of eDNA-based findings (DNA potentially being redistributed by e.g. water currents), the development of markers that allow a better coverage of all key taxonomic groups within the wide marine diversity, and the availability of sufficient and unambiguous DNA sequences in reference databases (Lacoursière-Roussel et al. 2018; Zaiko et al. 2018; Van der Loos and Nijland 2020). Reference databases are currently incomplete, sometimes lacking common taxa, including those already identified as potential or likely invaders. Further development of barcoding reference databases is therefore essential to support future monitoring and detection of NIS using molecular methods (Bylemans et al. 2019).
In this study, we developed a list of potential marine NIS for Svalbard waters, based on the published literature and records of NIS introductions elsewhere in the Arctic. The currently available DNA library was expanded by adding DNA barcodes for two marker regions (18S and COI) of 37 native and potential NIS for the Arctic. Furthermore, the eDNA method was applied for the first time in marine soft sediments of northwest Svalbard to assess the presence of native and potential marine NIS. This study provides insight into the effectiveness of this technique for monitoring NIS in the Arctic.
Material and methods
Preparing an overview of marine NIS for Svalbard
An initial list of 103 marine species recorded as potential NIS for the Arctic was derived from the list assembled on EMODnet’s SeaBasin Checkpoint—Arctic page on alien species (http://www.emodnet-arctic.eu/alien-species, access date 9 December 2020). This list includes a wide variety of groups ranging from phytoplankton to fish and plants. It was subsequently expanded with the marine NIS reported for Svalbard by Ware et al. (2016) and the Norwegian Biodiversity Information Centre (2018), as well as the marine NIS listed for the Arctic (mainly Barents Sea) by Chan et al. (2018).
Selection of barcode regions
The selection of barcode markers was based on an extensive screening for three complementary criteria: (1) the potential discrimination among taxa occurring in the North Sea and Arctic marine environment, (2) the availability of primers and protocols in peer-reviewed literature, and (3) the availability of reference data in online databases. Based on the first two criteria, 18S and COI were selected, being the most commonly used fragments and the only two barcodes for which sufficient reference data are available (Günter et al. 2018; Stefanni et al. 2018). The 18S functioned as a backbone, discriminating between key taxonomic groups among eukaryotes, and providing a general, wide screening of species. Despite its often lower taxonomic resolution, generic 18S markers have proven useful for genus and even species-level identifications in some groups including copepods (Wu et al. 2015). To obtain more information on the metazoa (animal kingdom) COI was used, enabling identification to species level because of the much higher resolution. The combination of 18S and COI therefore explored the added value of 18S and CO1 for NIS detection.
Specimen collection for generation of reference barcodes
The initial list of 103 NIS for the Arctic based on the EMODnet’s SeaBasin Checkpoint was used to identify key taxa with barcodes missing in online databases. Online barcoding libraries (e.g. Genbank, BOLD) were consulted for existing DNA barcoding markers (COI and 18S) for these Arctic NIS. From this screening, a total of 23 species were found to have full barcode coverage of the entire marker regions, 48 species had partial coverage (sequences covering e.g. only the V4 or V9 region of 18S) and 32 species had no coverage at all for COI and 18S markers (Online Resource 1). To generate both 18S and COI barcodes for the database for selected species with no (three species) or partial (nine species) coverage, specimens were collected from locations where the species were well established.
European specimens were collected by hand in various locations in the Oosterschelde in the Netherlands in August–September 2017 (Online Resource 1). Additionally, several Arctic specimens were collected from Kongsfjorden, northwest Svalbard, in July 2017, and barcoded to serve as positive controls of native species in the eDNA samples (Fig. 1). Each collected specimen was morphologically identified to species level, stored in 97% ethanol in a plastic tube, and transported to the Ecological Genetics Laboratory of Wageningen Environmental Research for DNA barcoding.
Generation of reference barcodes
DNA extraction from tissues (for Barcoding purposes) was performed using the DNeasy Blood & Tissue Kit of Qiagen according to the kit protocol.
Primers and PCR protocols were identical both for the reference barcoding and eDNA metabarcoding samples. For both markers (18S and CO1) we selected primer pairs that are most commonly used in the recent literature on eDNA metabarcoding of the marine realm. For 18S, primers were based on Stoeck et al. (2010), amplifying a ~ 270 bp fragment of the V4 region of the eukaryote SSU rRNA gene. We adopted their forward primer TAReuk454FWD1 while using an optimized version of their reverse primer TAReukREV3_(TAReukREV3_v1; 5′-ACTKTCGYTCWTGAYYRA-3′). Although the V9 region of 18S may have a higher taxonomic resolution and may therefore detect a higher diversity, a lack of reference data compared to V4 so far limits its potential for detailed taxonomic annotation of this diversity (see e.g. Choi & Park 2020). For COI, primers were based on Leray et al. (2013), amplifying a ~ bp fragment. Here, we adopted their reverse primer jgHCO2198, while using an optimized version of their forward primer mlCOIintF (mlCOIintF_v2; 5′-GGIACIGGITGRACWGTNTAYCCNCC-3′). Optimization consisted in both cases of creating a more degenerate version of the primer to increase amplification success for specific taxonomic groups (particularly Annelida, Arthropoda and Mollusca). In a direct comparison using the same PCR and bioinformatic protocols these versions detected a clearly higher taxon diversity than the original primers for the same samples (Glorius, Laros & De Groot, unpublished data). Forward and reverse primers were augmented with CS1 and CS2 tag sequences (Fluidigm, South San Francisco, CA, USA) to be used as sequence primers in Sanger sequencing (for reference barcodes, see below) and to allow the indexing PCR for multiplexed analysis via high-throughput sequencing (see below). PCR reactions were performed in a 25 µl reaction volume, consisting of 1U Platinum Taq (Fisher Scientific), 1 × PCR buffer, 2.5 mM MgCl2, 5%(m/m) Trehalose, 200 ng/µl BSA, 200 µM dNTP and 250 µM of each primer. The cycling program was as follows: 2 min at 94 °C followed by 15 cycles of 30 s at 94 °C, 3 min at 56 °C reduced by 1 °C each cycle and 1 min at 72 °C, followed by 20 additional cycles of 30 s at 94 °C, 3 min at 42 °C and 1 min at 72 °C and ended by a 10 min hold at 72 °C.
PCR products were then sent to an external company (Macrogen Europe B.V., Amsterdam, the Netherlands) for Sanger sequencing with both the forward and reverse primer per sample, and final consensus sequences per sample were determined by hand using Seqman (DNAStar).
Environmental sample collection
Samples for eDNA analysis were collected in four locations in Kongsfjorden, northwest Svalbard, in July 2017 (Fig. 1). A total of 30 marine sediment samples were collected using a Van Veen grab on-board the r/v Teisten: 26 in the Kongsfjorden (8 near Ny-Ålesund mine, 10 in Ny-Ålesund harbour, 8 near Lovénbreen) and 4 in the entrance of the Krossfjorden (see Online Resource 2). Plastic samples tubes (50 ml) were filled with approximately 40 g of sediment (wet weight) that was randomly collected from the upper 1 cm layer of the grab surface. The sample was kept cool until return to the Kings Bay Marine Laboratory at the end of each sampling day, where it was stored frozen (− 20 °C). Samples were transported in frozen condition to the laboratory in Wageningen for DNA metabarcoding.
eDNA metabarcoding
DNA extraction from sediments was performed using the Powermax soil DNA isolation kit (Mobio) with the following adaptation from the kit protocol: dispersing of the sediment samples by vortexing with beads was replaced by a dispersing using an Ultra Turrax (T25) instrument equipped with an in house constructed titanium dispersing shaft, while keeping the samples cooled on ice. This results in a better homogenization of the entire sample volume (see e.g. Aylagas et al. 2016). A subsample of 8 g of this homogenized volume was then included in the Powermax extraction.
PCR reactions were conducted following the above mentioned protocols for reference barcodes. The produced amplicons were then sent to Genome Quebec (Canada) for high-throughput sequencing. Here, a second PCR reaction was conducted to add sample-specific index barcodes and Illumina adaptor sequences. Indexed amplicons were then normalized and pooled per marker, before being sequenced in a 250 bp paired-end run on a Illumina Miseq flow cell platform.
Raw sequence data were processed in the R programming environment (R Core Team, 2017) making use of functions available in the DADA2 packages (Callahan et al. 2016). Primer sequences were removed from the raw sequence reads and the read quality was inspected by plotting the quality scores per base position for each sample. The following filtering steps were carried out; sequences with undetermined nucleotides, exceeding the expected number of errors of two and reads contaminated with the Phix genome were removed. All reads were trimmed at the point where read quality dropped below a score of two. The minimum sizes for the forward and reverse read lengths were set by inspecting the quality plots and varied between 202 and 230 depending on gene and forward/reverse read. The DADA 2 error model parameter learning algorithm was carried out on the sequences passing the filtering steps, followed by a dereplication step and inferring the sequence variance using standard settings of the DADA2 functions ‘derepFastq’ and ‘dada’. Forward and reverse reads were merged, and a table containing the number of reads per unique sequence variant per sample was constructed. In a final step, chimeras were identified and removed.
Automated taxonomic annotation of all obtained unique sequence variants was performed using a home-made script based on the BLAST® program, using the NCBI nucleotide database as a reference. To avoid false positive detections of NIS, we used a conservative approach for this automated annotation, assigning a taxonomy only to reads for which the top hits in BLAST had a sequence coverage of > 90% and a sequence similarity > 97% (following e.g. Holman et al. 2019). Reads were assigned to genus level in case all hits with a similarity > 97% belonged to the same genus. Likewise, reads were identified to species level in case the top hits had a similarity > 99% and all hits with a similarity > 99% belonged to the same species. Sequences that could not be identified at least to genus level were recorded as unidentified, as such results do rarely allow a reliable detection of NIS.
For all reads identified as potential NIS, we assessed the validity of this result via manual BLAST searches, analysing the diversity observed among the top 50 hits (based on similarity). For some reads assigned to genus level only, this allowed us to assess whether it more likely concerned a native species or a species reported as NIS for the Svalbard region, thereby discarding some reads as belonging to potential NIS.
Results
List of potential marine NIS for Svalbard
Based on all available information, a list of potential NIS was drafted for Svalbard marine waters consisting of 123 species. The list predominantly consists of arthropods (55%), along with chordates (14%), ochrophytes (7%), rhodophyte (5%), molluscs and nematodes (both 4%) species (Online Resource 3).
New reference barcodes
In total 37 species were barcoded; 31 of these were developed for the 18S marker, and 28 species for the COI marker (Online Resource 1). This included eight species identified as potential Arctic NIS and 13 Arctic species that acted as positive DNA references for the identification of species in the sediment samples. These barcodes were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/nucleotide). Successful barcodes could not be produced for the chordate Botryllus schlosseri, the chlorophyte Codium fragile, the ochrophyte Fucus serratus and the rhodophyte Caulacanthus okamurae/ustulatus (all collected from the Oosterschelde), as well as an unknown tube worm and Mya sp. from Kongsfjorden, Svalbard.
Metabarcoding of sediment samples
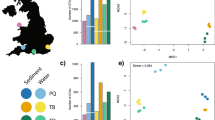
A total of 299 species were identified from the molecular analysis of the sediment samples, of which 243 were identified using the 18S marker and 64 using the COI marker. The following eight species were identified using both markers: the annelids Lysippe labiata, Scoloplos sp. and Terebellides stroemii; the ochrophytes Desmarestia sp. and Thalassiosira sp.; the chlorophyte Micromonas sp.; the mollusc Musculus niger; and the myzozoan Polarella glacialis. More species were detected in the sediment samples using the 18S marker (range of total number of species per location was 121–141) compared to the COI marker (range of total number of species per location was 28–33) (Table A-B in Online Resource 4). The phyla with the greatest number of species in the DNA reads obtained from the Kongsfjorden sediment samples using the 18S marker were nematods (18–30 species), followed by ochrophytes (17–20 species), chlorophytes (8–18 species), arthropods (8–13 species), and platyhelminthes (3–10 species) (Fig. 2a, Table A in Online Resource 4). The group ‘other’ comprises other, less common, taxa, including a wide diversity unicellular protists (see Online Resource 4). From the COI results, mainly ochrophytes (8–14 species) and annelids (7–13 species) were identified, followed by molluscs (2–4 species) (Fig. 2b, Table B in Online Resource 4). The COI results also included the identification of the terrestrial chironomid fly Micropsectra radialis, which was found despite it not being a marine species.
Of the 243 species identified using the 18S marker, 79 (33%) were identified to species level, and the remaining 164 to genus level. Of the 64 species identified using the COI marker 39 (61%) were identified to species level, and the remaining 25 to genus level.
Five of the newly barcoded Arctic species were positively identified in the sediment samples collected in Kongsfjorden, Svalbard. This included one species using the 18S marker (the chordate Pelonaia corrugata–5 samples) (Table A in Online Resource 4) and four species based on the COI marker (the molluscs Macoma calcarea–11 samples, Astarte borealis–2 samples, Mytilus sp.–1 sample, and the rhodophyte Polysiphonia arctica–7 samples) (Table B in Online Resource 4).
Identification of NIS
Based on our drafted list of potential NIS for Svalbard marine waters, seven potential NIS species were identified in the sediment samples collected in the Kongsfjorden; three using the 18S marker, and four using the COI marker (Table 1). The potential NIS consisted of three arthropods (copepods and a cirriped), one annelid (polychaete), two chordates (ascidians) and one mollusc (bivalve). Four were identified to species level: the copepods Euterpina acutifrons and Oithona similis, and the ascidians Botrylloides violaceus and Molgula manhattensis. Three were identified to a genus level: the polychaete Scolelepis sp., the cirriped Balanus sp. and the bivalve Mytilus sp.. Four were observed in a single sample: in the harbour (Mytilus sp., B. violaceus), near the mine (Balanus sp.), and near the Lovénbreen (M. manhattensis), whereas Scolelepis sp. and Euterpina acutifrons were found in 4 and 11 samples, respectively.
Discussion
Detection of NIS
eDNA metabarcoding shows great potential as an effective monitoring tool to assess the presence of NIS in environmental samples. This pilot study using sediment samples from Kongsfjorden, Svalbard, revealed seven species that are listed as potential Arctic NIS based on their DNA (Table 1). One of these is however the copepod O. similis, which is, although considered an NIS for Canadian ports (Brown et al. 2016), ubiquitous and widespread throughout the Arctic Basin and occurring naturally in Svalbard (Gulliksen et al. 1999; Ormanczyk et al. 2017). It should therefore be removed from the draft list of potential NIS for Svalbard.
Euterpina acutifrons is a harpacticoid copepod that was detected in 11 sediment samples in Kongsfjorden (based on the 18S marker). This may suggest that the species has established in the region, although its presence has not been reported in the Svalbard region before. It was identified earlier as an NIS in Canadian East Arctic-West Greenland and Hudson Bay, with vessels mentioned as a possible vector and with an unknown source region (Brown et al. 2016; Chan et al. 2018). No information is available on its potential impact on the ecology around Svalbard. In temperate and tropical areas E. acutifrons is a dominant member of some planktonic communities (Sautour and Castel 1993; Yahia et al. 2004; Vidjak et al. 2006). The dominance of the species is reliant on high concentrations of suspended particulate matter (Sautour and Castel 1993) and although E. acutifrons is reported to overwinter in the sediment at low temperatures (< 9 °C), breeding is limited in temperatures below 16 °C (D'Apolito and Stancyk 1979). Under current climate conditions the Arctic waters do not seem to meet the environmental parameters necessary for the species to become dominant.
The colonial chain tunicate B. violaceus and the solitary tunicate M. manhattensis (sea grape) were both identified in one sediment sample. Based on these data it cannot be concluded whether they were settled, or whether the positive identifications were based on alive or dead cells. Both tunicates are invasive in other regions and can cause significant negative impacts (Carver et al. 2006; Fofonoff et al. 2018). They can smother other species, and compete with filter feeders for food, potentially leading to serious changes in the local biodiversity (Carver et al. 2006; Carman et al. 2010). Economic problems of these species have been reported for aquaculture, fishing and shipping by clogging equipment and increasing maintenance costs (Carver et al. 2006; Fofonoff et al. 2018). Potential vectors of introduction to Svalbard are shipping and floating debris (Carver et al. 2006; Lejeusne et al. 2011). Under current climate conditions, B. violaceus can survive year-round in large areas of the Bering Sea shelf, but with limited opportunities for reproduction (Reimer et al. 2017). The temperature range for survival of this species lies between − 0.6 °C and 29 °C with a salinity range of 20–38 ‰, whereas for sexual reproduction this range was between 15 °C and 25 °C with a salinity of 26–38 ‰ (Reimer et al. 2017). Colony growth, however, can also occur by asexual budding and fragmentation which may enhance vegetative growth where established. Temperature survival of M. manhattensis is reported to be between − 1.5 °C and 32.1 °C with a mean salinity of 30.6‰ ± 5.6 (Lins et al. 2018). Both tunicates recently had first observations in southwestern Iceland in 2018, which is considered a first station for expansion of NIS towards very cold areas (Ramos-Esplá et al. 2020). The observed effects of both species in other invasion areas indicates the urgency to assess the route of introduction and establishment potential on Svalbard.
Another novel observation for Svalbard was that of the sabellid polychaete, Chone mollis, in one sediment sample of the Krossfjorden (based on the COI marker, Table 1). This species was not included in the list of potential NIS for Svalbard. It is known to occur in muddy sands of estuaries of the Pacific in the northern hemisphere (Bonar 1972), but has not been previously described for Svalbard. Related Chone spp. are however common to the Svalbard coast, such as Chone duneri, Chone infundibuliformis and Chone paucibranchiata (Gulliksen et al. 1999; Palerud et al. 2004). Further monitoring is needed to confirm whether Ch. mollis has established in the Svalbard region.
One sample in the harbour contained a positive identification for the mussel Mytilus sp.. Nowadays, several Mytilus spp. can be found on Svalbard: Mytilus edulis, Mytilus galloprovincialis or hybrids of M. edulis/M. galloprovincialis and M. edulis/Mytilus trossulus, although M. edulis is so far the most abundant species (Mathiesen et al. 2017; Węsławski and Kotwicki 2018). A recent study indicated that mussels (M. edulis) found on floating debris around Svalbard were hybrids of boreal and warm temperate taxa, some with genetic structure resembling populations in the UK, Iceland, and western mainland Europe (Wenne et al. 2020). This shows the complex interpretation of origin that may also account for other NIS.
Of the list of 123 potential NIS for Svalbard, 12 species have already been observed in earlier studies in this region (Online Resource 3): the annelid Spiophanes kroeyeri (Gulliksen et al. 1999; Palerud et al. 2004); the arthropods Crassicorophium bonellii, Lithodes maja and Paralithodes camtschaticus (Gulliksen et al. 1999; Palerud et al. 2004); the bryozoan Schizoporella unicornis (Gulliksen et al. 1999; Palerud et al. 2004); the chordates Botryllus schlosseri and Ciona intestinalis (Gulliksen et al. 1999; Palerud et al. 2004); the echinoderm Echinus esculentus (Gulliksen et al. 1999; Palerud et al. 2004); the mollusc Mya arenaria (Gulliksen et al. 1999; Palerud et al. 2004); the ochrophyte Fucus serratus (Gulliksen et al. 1999), and the rhodophyte Dumontia contorta (Gulliksen et al. 1999). Whether all these species were correctly labelled as potential NIS to Svalbard needs further study. The annelid Heteromastus filiformis, also listed as potential NIS for Svalbard and observed in earlier studies, is not considered an NIS to Svalbard. Another 26 species of the list of potential NIS for Svalbard have been identified in ballast water samples collected from ships arriving to Svalbard (Ware et al. 2016) (Online Resource 3).
It is important to realise that not all newly arriving marine species become invasive or form a nuisance to the receiving habitat. A newly arrived NIS may settle, grow and reproduce, while slowly obtaining an ecological niche in the new habitat. With the warming waters in the Arctic region, the coastal waters around Svalbard are expected to become more hospitable to more southern species in the near future (Ware et al. 2016), while indigenous species may be less adapted to the new conditions. Although only a few of the many introduced species may become pests, the impact of these may be large (Mack et al. 2000). Therefore, it is better to prevent anthropogenic introductions of new species, as measures for prevention are more realistic than measures for removal of already established NIS.
Using eDNA metabarcoding as a monitoring tool
Some of the potential NIS found in the sediment samples could only be identified at a higher taxonomic level than specified in the list of potential Arctic NIS (Table 1). Based on genus level, no confirmation on the presence of the actual Arctic NIS can be made as many of the taxa identified to genus level may be species that are native to Svalbard, such as e.g. Balanus balanus, Mytilus edulis, Scolelepis burkovskii, and Scolelepis squamata (Gulliksen et al. 1999; Varpe 2012; Sikorski and Pavlova 2015). Incomplete identification of species in the sediment samples was generally due to the absence in the references library of either the native or the invasive species, and/or the 18S and COI marker regions showing insufficient polymorphisms between species within a genus to properly distinguish them.
This further stresses the need for the publication of species-specific barcodes for commonly used DNA markers (such as 18S and COI) in international reference databases, such as the online BOLD and NCBI databases. This will ultimately lead to a more detailed identification so that NIS sharing a genus with native species can be successfully distinguished. In this study, we provided new barcodes for 31 marine species, of which eight are listed as potential Arctic NIS. Unfortunately, barcodes for 14 of the 123 species from the list of potential marine NIS for Svalbard, are still missing for both the 18S and COI marker (Online Resource 5). An additional 33 have missing barcodes for either the 18S or the COI marker. A follow-up project to update the international barcode databases for these species is therefore essential to enable a more complete identification of marine NIS on Svalbard.
This study was a proof-of-principle for using sediments for the identification of NIS. These data can however also be used for assessing species diversity. Already in the relatively small sample volume (40 g ww of the top 1 cm), 299 species were revealed. For a proper baseline description of biodiversity, of course, an optimised number of samples is needed per sampling site. In our study we found that for the COI marker up to seven to eight sediment samples were needed to obtain 95% of the total observed species at NA harbour, NA mine and the Lovénbreen (Figure A in Online Resource 6). For the 18S marker, up to ten samples were needed to obtain 95% of the observed species at the sampling sites. As the species-accumulation curves from all sampling sites for this marker had not reached assymptotes yet, this indicates the ideal number of samples will be more than ten (Figure B in Online Resource 6). Care should be taken to use these minimum sample numbers as a fixed number of samples for all sites. Not all species in the environment are currently covered by the molecular markers and site heterogeneity also influences the required number of samples. For marine water samples it was suggested to collect at least 15 samples for a comprehensive estimate of biodiversity at a site (Lacoursière-Roussel et al. 2018).
A combination of different sample types will further complement the information on the presence of marine species (including NIS) in a particular area, such as water samples for mobile species, hard substrate scrapes or plastic debris. For example, we identified 26 species on a piece of floating soft plastic in Ny-Ålesund harbour that were not found in the harbour sediment (see Online Resource 7). Floating debris can act as rafts onto which species can attach and travel significant distances on ocean currents (Carlton et al. 2017). Encrusting species such as the bryozoans Electra spp. and Eucratea loricata, the cirripeds Semibalanus balanoides and Lepas anatifera, and the bivalve Mytilus sp.were previously observed on large plastics items such as boxes, barrels and liquid containers that were washed upon beaches of Prins Karl Forlandet, Svalbard (Węsławski and Kotwicki 2018), with L. anatifera being not native to Svalbard. The considerable amount of floating plastic in the oceans increases the risk of introducing hitchhiking NIS. Coastal monitoring efforts should therefore also include floating plastic as a potential vector of NIS introduction.
At present the identification of species in eDNA may not provide direct information on whether the species is alive or not. A positive identification however shows that DNA fragments were present at the monitoring site. For screening purposes of NIS therefore, this information forms an important first step for selecting sites for further analysis and applying a more thorough study using visual inspection.
Future perspectives
Metabarcoding is a suitable technique for rapid detection of the presence of species in coastal ecosystems. It enables analysing samples en masse without the expertise-intensive taxonomic identification of individual specimens. The 18S and COI markers provided complementary information, in total identifying 299 species in the sediment samples, with only eight species overlapping for both markers. Many of the identified species came from groups of microorganisms and from taxa that are rarely studied and sampled in this region, such as flatworms, fungi, slime molds, and parasites from a variety of phyla, especially using the 18S marker as general wide screening of species (Tables A and B in Online Resource 4). This shows that the technique has great potential for describing otherwise understudied local biodiversity. This also accounts for early life stages and organisms that are hard to detect by visual taxonomic identification techniques, and can assist by rapidly enhancing information on the occurrence of such species.
Already the limited sampling performed in this study indicated the presence of marine NIS on Svalbard and showed that the eDNA metabarcoding method is well-suited for the detection and monitoring of the presence of NIS. The novel observations of E. acutifrons, B. violaceus, M. manhattensis and Ch. mollis in our pilot study warrant further detailed studies to assess whether these species established in Svalbard waters and to unravel their vector of introduction and point of arrival, as well as their potential impact on the system. Passenger and cargo shipping is likely the primary vector of marine NIS to Svalbard, whereby they can facilitate NIS introduction through hull fouling, ballast waters and discards of food products (primarily from cruise ships). In addition, floating debris can serve as both long and short distance vectors, whereas natural spreading can occur from NIS introduced to mainland Norway in recent years. We argue that the likely points of arrival via most of these vectors are in areas such as harbours (Longyearbyen, Pyramiden, Barentsburg, Svea and Ny-Ålesund), exposed shorelines and places where drifting marine litter is piling up on beaches. In fact, our focus on the Kongsfjorden system (Ny-Ålesund) likely gives a very conservative picture of the presence of marine NIS on Svalbard as the Isfjorden (Longyearbyen and Barentsburg) experiences much higher ship traffic and the exposed west coasts of Spitsbergen north of Van Mijenfjord are hotspots of drifting debris.
Monitoring the introduction vectors and presence of NIS in Svalbard waters can lead to effective mitigation measures to stop the further introduction and anthropogenic spreading of NIS. We suggest to focus on strategic points of entry, such as those mentioned above, during the busy traffic months in summer. Frequent surveillance of ship hull fouling, and ballast water (see Ware et al. 2016), combined with targeted sampling of harbour and coastal habitats and education of local residents to identify likely NIS, are also advised to increase effectiveness in monitoring. A similar monitoring strategy is advisable to other harbours in the Arctic region as information on marine NIS is urgently lacking for this region. Finally, ecological studies of the potential impacts of these organisms are needed to assess whether the negative effects on the ecosystem, as observed elsewhere, may also account for Svalbard.
References
Aylagas E, Mendibil I, Borja Á, Rodríguez-Ezpeleta N (2016) Marine sediment sample pre-processing for macroinvertebrates metabarcoding: mechanical enrichment and homogenization. Front Mar Sci 3:203
Bax N, Williamson A, Aguero M, Gonzalez E, Geeves W (2003) Marine invasive alien species: a threat to global biodiversity. Mar Policy 27:313–323
Berge J, Johnsen G, Nilsen F, Gulliksen B, Slagstad D (2005) Ocean temperature oscillations enable reappearance of blue mussels Mytilus edulis in Svalbard after a 1000 year absence. Mar Ecol Prog Ser 303:167–175
Berge J, Renaud P, Bluhm B, Danielsen H (2017) Om snøkrabbe, torsk og sjømat fra havet rundt Svalbard. Svalbardposten 25(08):2017
Bonar DB (1972) Feeding and tube construction in Chone mollis Bush (Polychaeta, Sabellidae). J Exp Mar Biol Ecol 9:1–18
Brown EA, Chain FJJ, Zhan A, MacIsaac HJ, Cristecu ME (2016) Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Divers Distrib 22:1045–1059
Buchholz F, Buchholz C, Węsławski JM (2010) Ten years after: krill as indicator of changes in the macro-zooplankton communities of two Arctic fjords. Polar Biol 33:101–113
Bylemans J, Gleeson DM, Duncan RP, Hardy CM, Furlan EM (2019) A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environ DNA 1:402–414
CAFF (2013) Arctic Biodiversity Assessment. Status and trends in Arctic biodiversity, Conservation of Arctic Flora and Fauna, Akureyri
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Carlton JT, Chapman JW, Geller JB, Miller JA, Carlton DA, McCuller MI, Treneman NC, Steves BP, Ruiz GM (2017) Tsunami-driven rafting: Transoceanic species dispersal and implications for marine biogeography. Science 357:1402–1406
Carman MR, Morris JA, Karney RC, Grunden DW (2010) An initial assessment of native and invasive tunicates in shellfish aquaculture of the North American east coast. J Appl Ichthyol 26:8–11
Carver CE, Mallet AL, Vercaemer B (2006) Biological Synopsis of the colonial tunicates, Botryllus schlosseri and Botrylloides violaceus. Canadian Manuscript Report of Fisheries and Aquatic Sciences 2747: v + 42p
Chan FT, Stanislawczyk K, Sneekes AC, Dvoretsky A, Gollasch S, Minchin D, David M, Jelmert A, Albretsen J, Bailey SA (2018) Climate change opens new frontiers for marine species in the Arctic: Current trends and future invasion risks. Glob Change Biol 25:25–38
D’Apolito LM, Stancyk SE (1979) Population dynamics of Euterpina acutifrons (Copepoda: Harpacticoida) from North Inlet, South Carolina, with reference to Dimorphic Males. Mar Biol 54:251–260
Deja K, Węsławski JM, Borszcz T, Włodarska-Kowalczuk M, Kukliński P, Bałazy P, Kwiatkowska P (2016) Recent distribution of Echinodermata species in Spitsbergen coastal waters. Pol Polar Res 37:511–526
Fofonoff PW, Ruiz GM, Steves B, Simkanin C, Carlton JT (2018) National Exotic Marine and Estuarine Species Information System. http://invasions.si.edu/nemesis/.Access Date: 10-Dec -2020
Førland EJ, Benestad R, Hanssen-Bauer I, Haugen JE, Skaugen TE (2011) Temperature and precipitation development at Svalbard 1900–2100. Advances in Meteorology. https://doi.org/10.1155/2011/893790
Goldsmit J, Howland KL, Archambault P (2014) Establishing a baseline for early detection of non-indigenous species in ports of the Canadian Arctic. Aquat Invasions 9:327–342
Gulliksen B, Palerud R, Brattegard T, Sneli J (1999) Distribution of marine benthic macro-organisms at Svalbard (including Bear Island) and Jan Mayen. Research report for DN 1999–4 Directorate for Nature Management.
Günter B, Knebelsberger T, Neumann H, Laakmann S, Martinez Arbizu P (2018) Metabarcoding of marine environmental DNA based on mitochondrial and nuclear genes. Sci Rep 8:14822
Holman LE, De Bruyn M, Creer S, Carvalho G, Robidart J, Rius M (2019) Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci Rep 9:11559
Jeunen GJ, Knapp M, Spencer HG, Lamare MD, Taylor HR, Stat M, Gemmell NJ (2019) Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol Ecol Resour 19:426–438
Jørgensen LL, Manushin I, Sundet JH, Birkely S-R (2005) The intentional introduction of the marine red king crab Paralithodes camtschaticus into the Southern Barents Sea. ICES Cooperative Research Report No. 277
Lacoursière-Roussel A, Howland K, Normandeau E, Grey EK, Archambault P, Deiner K, Lodge DM, Hernandez C, Leduc N, Bernatchez L (2018) eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol Evol 8:7763–7777
Lejeusne C, Bock DG, Therriault TW, MacIsaac HJ, Cristescu ME (2011) Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol Invasions 13:635–650
Leopold P, Renaud PE, Ambrose WG, Berge J (2019) High Arctic Mytilus spp.: occurrence, distribution and history of dispersal. Polar Biol 42:237–244
Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34
Lins DM, de Marco JP, Andrade AF, Rocha RM (2018) Predicting global ascidian invasions. Divers Distrib 24:692–704
Mack RN, Simberloff D, Mark Lonsdale W, Evans H, Clout M, Bazzaz FA (2000) Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl 10:689–710
Mathiesen SS, Thyrring J, Hemmer-Hansen J, Berge J, Sukhotin A, Leopold P, Bekaert M, Sejr MK, Nielsen EE (2017) Genetic diversity and connectivity within Mytilus spp. in the subarctic and Arctic. Evol Appl 10:39–55
Molnar JL, Gamboa RL, Revenga C, Spalding MD (2008) Assessing the global threat of invasive species to marine biodiversity. Ecol Environ 6:485–492
Norwegian Biodiversity Information Centre (2018). The alien species list of Norway – ecological risk assessment 2018. Retrieved in January 2019 from https://www.biodiversity.no/alien-species-2018
Ormanczyk MR, Gluchowska M, Olszewska A, Kwasniewski S (2017) Zooplankton structure in high latitude fjords with contrasting oceanography (Hornsund and Kongsfjorden, Spitsbergen). Oceanologia 59:508–524
Palerud R, Gulliksen B, Brattegard T, Sneli JA, Vader W (2004) The marine macro-organisms in Svalbard waters. In: Strøm H, Prestrud P, Goldman HV (eds) A catalogue of the terrestrial and marine animals of Svalbard. Norwegian Polar Institute, Norway
Ramos-Esplá AA, Micael J, Halldórsson HP, Gíslason S (2020) Iceland: a laboratory for non-indigenous ascidians. Bioinvasions Rec 9:450–460
Reimer JP, Droghini A, Fischbach A, Watson JT, Bernard B, Poe A (2017) Assessing the Risk of Non-Native Marine Species in the Bering Sea. NPRB Project 1523. Alaska Center for Conservation Science, University of Alaska Anchorage, p 39
Ruiz GM, Hewitt CL (2009) Latitudinal patterns of biological invasions in marine ecosystems: A polar perspective. Smithsonian Institution Scholarly Press, Washington DC, pp 347–358
Ruppert KM, Kline RJ, Rahman MS (2019) Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Eco Cons 17:e00547
Sautour B, Castel J (1993) Feeding behaviour of the coastal copepod Euterpina acutifrons on small particles. Cah Biol Mar 34:239–251
Sikorski A, Pavlova L (2015) New species of Scolelepis (Polychaeta, Spionidae) from the Norwegian coast and Barents Sea with a brief review of the genus. Fauna Norvegica 35:9–19. https://doi.org/10.5324/fn.v35i0.1666
Simberloff D, Parker IM, Windle PN (2005) Introduced species policy, management, and future research needs. Front Ecol Environ 3:12–20
Stefanni S, Stankovic D, Borme D, De Olazabal A, Juretic T, Pallavicini A, Tirelli V (2018) Multi-marker metabarcoding approach to study mesozooplankton at basin scale. Sci Rep 8:12085
Stoeck T, Bass D, Nebel M, Christen R, Jones MD, Breiner HW, Richards TA (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19:21–31
Trebitz AS, Hoffman JC, Darling JA, Pilgrim EM, Kelly JR, Brown EA, Chadderton WL, Egan SP, Grey EK, Hashsham SA, Klymus KE, Mahon AR, Ram JL, Schultz MT, Stepien CA, Schardt JC (2017) Early detection monitoring for aquatic non-indigenous species: Optimizing surveillance, incorporating advanced technologies, and identifying research needs. J Environ Manage 202:299–310
Van der Loos LM, Nijland R (2020) Biases in bulk: DNA metabarcoding of marine communities and the methodology involved. Mol Ecol 00:1–19
Varpe Ø (2012) Fitness and phenology: annual routines and zooplankton adaptations to seasonal cycles. J Plankton Res 34:267–276
Vidjak O, Bojanić N, Kušpilić G, Marasović I, Gladan Ž, Brautović I (2006) Annual variability and trophic relations of the mesozooplankton community in the eutrophicated coastal area (Vranjic Basin, eastern Adriatic Sea). J Mar Biol Assoc UK 86:19–26
Walther GR (2009) Alien species in a warmer world: risks and opportunities. Trends Ecol Evol 24:686–693
Ware C, Berge J, Jelmert A, Olsen SM, Pellissier L, Wisz M, Kriticos D, Semenov G, Kwasniewski AIG (2016) Biological introduction risks from shipping in a warming Arctic. J Appl Ecol 53:340–349
Wenne R, Zbawicka M, Bach L, Strelkov P, Gantsevich M, Kuklinski P, Kijewski T, McDonald JH, Sundsaasen KK, Arnyasi M, Lien S, Kaasik A, Herkul K, Kotta J (2020) Trans-Atlantic Distribution and Introgression as Inferred from Single Nucleotide Polymorphism: Mussels Mytilus and Environmental Factors. Genes 11:530
Węsławski JM, Kotwicki L (2018) Macro-plastic litter, a new vector for boreal species dispersal on Svalbard. Pol Polar Res 39:165–174
Węsławski JM, Kendall MA, Włodarska-Kowalczuk M, Iken K, Kędra M, Legezynska J, Sejr MK (2011) Climate change effects on Arctic fjord and coastal macrobenthic diversity – observations and predictions. Mar Biodivers 41:71–85
Węsławski JM, Dragańska-Deja K, Legeżyńska J, Walczowski W (2018) Range extension of a boreal amphipod Gammarus oceanicus in the warming Arctic. Ecol Evol 8:7624–7632
Witkowski A, Glowacki P (2010) A record of pink salmon, Oncorhynchus gorbuscha (Actinopterygii, Salmoniformes, Salmonidae), in the Revelva River, Hornsund area (SW Spitsbergen). Acta Ichthyol Piscat 40:87
Wu S, Xiong J, Yu Y (2015) Taxonomic Resolutions Based on 18S rRNA Genes: A Case Study of Subclass Copepoda. PLoS ONE 10:e0131498
Yahia M, Souissi S, Yahia-Kéfi OD (2004) Spatial and Temporal Structure of Planktonic Copepods in the Bay of Tunis (Southwestern Mediterranean Sea). Zool Stud 43:366–375
Zaiko A, Pochon X, Garcia-Vazquez E, Olenin S, Wood SA (2018) Advantages and Limitations of Environmental DNA/RNA Tools for Marine Biosecurity: Management and Surveillance of Non-indigenous Species. Front Mar Sci 5:322
Acknowledgements
This work was supported by the Svalbard Environmental Protection Fund (17/25-Marine aliens in Svalbard), the Knowledge Base Programme of Wageningen University and Research (431.830.0049), and the Norwegian Environmental Agency (Coastal biodiversity and introduced species in Arctic Norway, 18087179). The Governor of Svalbard provided sampling permission. Frits Steenhuisen, Michiel Klaassen, Mare van den Heuvel, Maarten Loonen (Netherlands Arctic Station), Svein Harald Sønderland (Ny-Ålesund Marine Laboratory), Roar Strand (Teisten), Bart van den Heuvel, Nanne van den Heuvel and Mario de Kluijver are thanked for their assistance during field work. The reviewers are kindly acknowledged for their constructive suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van den Heuvel-Greve, M.J., van den Brink, A.M., Glorius, S.T. et al. Early detection of marine non-indigenous species on Svalbard by DNA metabarcoding of sediment. Polar Biol 44, 653–665 (2021). https://doi.org/10.1007/s00300-021-02822-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-021-02822-7