Abstract
Glacier forelands are excellent sites in which to study microbial succession because conditions change rapidly in the emerging soil. Development of the bacterial community was studied along two transects on lateral moraines of Ecology Glacier, King George Island, by culture-dependent and culture-independent approaches (denaturating gradient gel electrophoresis). Environmental conditions such as cryoturbation and soil composition affected both abundance and phylogenetic diversity of bacterial communities. Microbiocenosis structure along transect 1 (severe cryoturbation) differed markedly from that along transect 2 (minor cryoturbation). Soil physical and chemical factors changed along the chronosequence (time since exposure) and influenced the taxonomic diversity of cultivated bacteria, particularly along transect 2. Arthrobacter spp. played a pioneer role and were present in all soil samples, but were most abundant along transect 1. Cultivated bacteria isolated from transect 2 were taxonomically more diverse than those cultivated from transect 1; those from transect 1 tended to express a broader range of enzyme and assimilation activities. Our data suggest that cryoturbation is a major factor in controlling bacterial community development in postglacial soils, shed light on microbial succession in glacier forelands, and add a new parameter to models that describe succession phenomena.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glacial retreat is one of the most conspicuous signs of warming in polar regions. The resulting ice-free terrain offers an excellent opportunity for ecological research (Schmalenberger and Noll 2010). Deglaciated terrain presents a chronosequence of soil formation, primary succession, and ecosystem development. Distance from a retreating glacier reflects soil age, with vegetation-free sandy deposits at the front of the glacier contrasting with more differentiated soils further away (Schmidt et al. 2008). Antarctic soils that only recently became ice-free are considered to be nutrient limited (Sigler et al. 2002; Darmody et al. 2005), despite the influence of local meltwater streams, organic and inorganic inputs from marine aerosols, guano deposits in penguin and other seabird rookeries, slow-growing cryptogamic plants, and marine algae deposited on the shore and dispersed by wind (Orchard and Corderoy 1983; Tibbles and Harris 1996; Beyer et al. 1999; Tockner et al. 2002; Grzesiak et al. 2009). Therefore, an urgent need exists to understand nutrient- and carbon-cycling processes in these recent and emerging environments.
Microorganisms are important in nutrient cycling in postglacial environments (Tscherko et al. 2003b) but are negatively affected by long periods of snow cover, low temperatures, and accompanying low water availability (Bardgett et al. 2007). Other studies, however, indicate that C and N pools increase in the first 20 years of succession, hinting at C and N fixation in these soils (Nemergut et al. 2007). The prevailing view is that photoautotrophs such as algae, mosses, lichens, and higher plants start primary succession on newly exposed surfaces by fixing carbon and promoting the accumulation of organic material necessary for development of heterotrophic communities (Walker and del Moral 2003). However, Tscherko et al. (2003a) show that heterotrophic microbial communities occur in newly exposed glacial terrain before photoautotrophic communities establish. Heterotrophic microbes clearly play an important role in the progress of primary succession in nutrient-limited soils in polar regions.
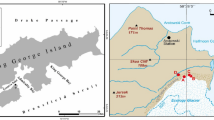
The main focus of the research described here is deglaciated soils in the vicinity of Ecology Glacier (King George Island, South Shetland Islands, Antarctica), whose retreat between 1978 and 2007 equates to 16.4 % (0.58 km2) of its 1978 surface area (Pudełko, Potocki, Angiel unpublished). Ecology Glacier moraines are influenced by cryoturbation and solifluction (an erosion process) (Blume et al. 2002a). Cryoturbation mainly occurs during the summer months when there is no snow on the soil surface, and the process is primarily based on the freeze–thaw cycle of water in the soil. Rock and mineral fragments break during these cycles (Blume et al. 2002b). The intensity of cryoturbation is evident in the fine silt and clay content of the soils (Seppelt 2002). During solifluction, thawed soil and rock move slowly downhill. Moreover, solifluction and cryoturbation hamper the establishment of permanent extended vegetation cover (Bölter and Blume 2002). High silt contents are also unfavorable for roots because intensive cryoturbation prevents stabilization of the soil fabric. Thus, silty to loamy soils affected by active cryoturbation lack plant cover (Beyer et al. 2002). Conditions in these soils may also change over short distances, and in many cases over meter scales. Mosses or lichens may develop in places where cryoturbation is absent, especially on sandy soils with gravel (Beyer et al. 2002). Frost impact is then reduced by a thick plant cover (Blume et al. 2002b).
Plant communities in ice-free areas around Ecology Glacier have been studied extensively (Olech 2002; Smykla et al. 2007; Parnikoza et al. 2008; Victoria et al. 2009), but little is known about bacterial succession, especially when there is ongoing cryoturbation. We hypothesize that varying soil conditions here have a large discriminating effect on the nature of the microbial community, leading to the establishment of dominant and robust ‘pioneer’ microbes. We thus investigated microbial community development on Ecology Glacier moraines, focusing especially on culturable heterotrophic bacteria in terms of their abundance, diversity, and morphophysiology.
Materials and methods
Sites and sampling
Surface soil samples (~0.5 kg) were collected in the vicinity of Ecology Glacier into sterile plastic bags using sterile plastic spatulas, in the 2008/2009 austral summer. One sample was also taken from Ecology Glacier itself as ice cores of the glacier’s surface with an auger (20 cm long, 2.5 cm diameter) (Table 1); pooled cores totaled ~0.5 kg and were allowed to melt in a refrigerator (~4 °C) before being processed for microbiological analyses. A duplicate set of cores was not thawed, but was stored at −20 °C in order to provide material for further research in Poland.
Soil at different distances from Ecology Glacier was collected in areas with less (transect 1, termed TR1) and more (transect 2, termed TR2) stable ground (Table 1): TR1, closer to the island’s rock massifs, lacks vegetation (samples collected here are termed 0, and A1 to E1); TR2, hosts plant communities of varying complexity (samples collected here are termed A2 to F2). Time since exposure of the ground along these transects was estimated by reference to maps (Birkenmajer 2002; Pudełko 2003, 2008); samples with the same letter names (e.g., A1 and A2) from both transects were exposed around the same time. Consecutive letters were given to samples collected at increasing distances from the glacier. Three ‘zones’ were identified: Zone I, exposed by glacial retreat in the last 5 years (samples 0, A1, A2); Zone II, exposed since 1978 and comprising mainly ground moraines almost devoid of vegetation cover, particularly along TR1, while terrain along TR2 hosts mosses (samples B1, B2, C1, C2); Zone III, comprising neoglacial moraines with more mature vegetation and the commonly occurring Usnea, indicative of an age of dozens to several 100 years (samples D1, D2, E1, E2, F2). Samples for TR1 in this zone were gathered from spots of ground that lacked vegetation, indicating cryoturbation influence.
Measurement of soil components
Granulometric composition of the soil was determined by sieving, according to Polish Standards (PN-R-04033: 1998). Soil matrix (stone/gravel, sand and silt/clay) fractions were separated by sieve (2 mm mesh) and 0.05-mm mesh. Each fraction was weighed to determine percentage contribution. The pH of 1 g (wet wt.) of soil suspended in 100 ml sterile distilled water was determined with a Hanna Instruments HI 9025 pH meter; wet weight was determined directly after sampling. Dry weight was determined after 24-h incubation at 65 °C in a dry box with circulating air (Dowgiałło 1975). Dried soil was powdered and used for elementary CHN analysis (Carlo Erba CHNS-O-meter, EA 1108). Phosphorus was determined in dried material by the colorimetric meta-vanadate method, and other elements were determined by atomic absorption spectroscopy (Tatur et al. 1997). Ash content was obtained by weighing the residue after combustion at 450 °C. The mean and standard errors (SEs) in water content, dry and wet weights, were determined from three measurements.
Bacterial abundance
Bacterial counts were determined in 1–2 g wet wt. soil and melted glacier ice. Soil was suspended in 20 ml sterile saline (0.85 %) in 100-ml sterile flasks with glass beads and shaken gently on a universal shaker (type 327) (120 rpm, 20 min, 5 °C). Suspensions were then stored in the refrigerator (4 °C) for 10–20 min to allow larger particles to settle. A decimal dilution series of the supernatant to 10−5 was prepared in sterile 0.85 % saline.
Sub-samples for total counts (TCs) were fixed with buffered formalin to a final concentration of 1 %. TCs were determined by epifluorescence microscopy using 4′,6-diamidino-2-phenylindole (DAPI) on black Nuclepore polycarbonate 0.2-μm-pore-size filters (Porter and Feig 1980), under a Nikon E-200 microscope with a 100 W Hg lamp and 100× CFI 60 oil immersion objective, with a digital DS Cooled Camera Head DS-5Mc-U1, and a filter block of wavelengths EX 330-380, DM 400, BA 420. Images of fields were analyzed in Nikon NIS Elements BR 2.30 and MultiScan v. 14.02 (computer scanning systems). A minimum of 400 cells in 20 fields per sample were counted automatically in the image analysis system. Cell volume and total bacterial biomass were evaluated through image analysis (Sieracki et al. 1985; Świątecki 1997). Cell volume (V) was converted to bacterial biomass in terms of carbon (C), where C (fg) = 120 × V 0.72 (Simon and Azam 1989). Average values of three measurements using three independently prepared filters were calculated.
Photoautotrophs, including cyanobacteria and photoautotrophic eukaryotes, were counted under blue 450–490 nm (B-2A Nikon filter) and green 510–560 nm (G-2A Nikon filter) light excitation (Putland and Rivkin 1999). Cyanobacteria were distinguished from photoautotrophic eukaryotes because of the former’s gold-yellow autofluorescence (Rassoulzadegan and Sheldon 1986).
A cell morph type diversity analysis describing the frequency of morphological forms in each size class, based on cell shapes (cocci, rods, curved forms) in five classes of volumes (<0.1; 0.1–0.2; 0.2–0.5, 0.5–1.0; >1 μm3), was conducted with a modified Shannon index (Gurienowich 1995; Nübel et al. 1999).
Denaturating gradient gel electrophoresis analysis
The bacterial community in each sample was compared through denaturating gradient gel electrophoresis (DGGE). DNA was extracted from 10 g of each soil sample using the MegaSoil DNA isolation Kit (MoBio, USA), according to the manufacturer’s protocol. The concentration of the extracted DNA was determined by measuring absorbance at 260 nm, and purity was estimated from the A 260/A 280 ratio. The V2–V5 regions in the 16S rRNA gene in the template DNA were amplified by polymerase chain reaction (PCR) with the primer 341f with a GC clamp 5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GCC TAC GGG AGG CAG CAG-3′, complementary to position 341–357, and 907r (CCG TCA ATT CMT TTG AGT TT) complementary to positions 926–907 (Escherichia coli numbering) (Crump et al. 2003, 2007). PCRs were performed in an MJ Mini™ Gradient Thermal Cycler (BioRad Laboratories, USA). The PCR (50 μl) contained 1× PCR buffer, 200 ng bovine serum albumin (Sigma-Aldrich Co.), 2.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 0.25 μM (each) primer, 1.5 U Taq polymerase (Sigma-Aldrich Co.) and approximately 1–2 ng μl−1 template DNA. PCR conditions were as follows: initial denaturation (94 °C, 5 min), followed by 10 touchdown cycles (denaturation 94 °C/1 min, annealing 65 °C (decrease 1 °C/cycle) and elongation 72 °C/3 min), 20 standard cycles (94 °C/1 min, 55 °C/1 min, 72 °C/5 min); the final elongation step was 5 min. Amplification products were visualized in a 1 % (wt/vol) agarose gel stained with ethidium bromide. Amplified DNA was separated using the D-Code Universal Mutation Detection System (BioRad Laboratories, USA) DGGE with a 6 % (wt/vol) 16 × 16 cm polyacrylamide gel (37:1 acrylamide:bisacrylamide) in 1× TAE buffer (40 mM tris, 20 mM acetic acid, 1 mM EDTA, pH 8.0) with a 35–70 % denaturant gradient (where 100 % denaturant was 7 M urea and 40 % formamide). Electrophoresis was at constant voltage (60 V) for 17 h at 60 °C. Electrophoresis products were stained by gently agitating the gel for 30 min in 100 ml 1× TAE containing 5 μl of a 1:10,000 dilution of SYBR Gold nucleic acid stain (Molecular Probes Inc., PoortGebouw, The Netherlands). DGGE gel images were analyzed using the Quantity One software on a GelDoc gel documentation system (BioRad Laboratories, USA). Gel bands were identified using GelCompar software to create a presence–absence matrix (Crump and Hobbie 2005). Each band represents a bacterial OTU (operational taxonomic unit).
Diversity of cultivated bacteria
Colony-forming units (CFU) enumerated by in a decimal dilution series of the sample suspension by the spread plate method on soil extract agar (SEA) were considered copiotrophic bacteria (CBcop), while those on 100× diluted SEA were considered oligotrophic bacteria (CBoligo) (Ogram and Feng 1997). The composition of SEA, per liter, is bacto beef extract (3 g), bacto-tryptone (5 g), bacto-agar (15 g) (Fenchel and Hemmingsen 1974) in 1 l of soil extract, prepared by autoclaving 200 g of Antarctic soil in 1 l water (Klement et al. 1990; Zdanowski et al. 2005). Inoculated plates were incubated in darkness at 4 °C and inspected every second to third day for 30 days for colony development and growth; this also enabled observation of particular colony types, and the rate at which CFU grew, from different sampling points on the moraine. CFU numbers either did not increase or increased only minimally, after 30 days. In that respect, CFU numbers by 7 days of incubation (considered fast-growing isolates) were compared with those determined periodically up to the 30th day of incubation (considered slow-growing isolates). CFU abundance was calculated on the basis of the number of colonies present at the end of the incubation period, with the SE being determined from three replicates.
After 30 days of incubation, several colony types that differed in terms of size, color, shape, and other colony characteristics were selected per sample. The number selected per sample varied depending on the diversity. Colonies of a particular type were counted and their number considered during morphophysiological tests and taxonomic identification. Pure isolates derived from streaking colonies for isolation were passed through morphophysiological tests, including Gram reaction, pigment production, ability to grow at 4, 22, and 32 °C on a nutrient-rich medium, and responses in the API 20NE and API ZYM test kits. A total of 46 morphological physiological and biochemical tests were performed on each isolate. The following tests were conducted: ability to grow at 4, 22, 32 °C; morphological analyses, which comprised counts of pigmented colonies (PIGM), two microscopy tests—Gram reaction (GRAM), morphology (ROD); the API 20NE system which allows nine biochemical tests: reduction in nitrate to nitrite (NO3), and to nitrogen (N2); indole production (TRP); fermentation of glucose (GLU_F); arginine dihydrolase (ADH); urease ((URE); β-glucosidase (ESC); gelatin hydrolysis (GEL); cytochrome oxidase (OX); and 12 tests for assimilation of carbohydrates as sole carbon sources; glucose (GLU); arabinose (ARA), mannose (MNE); mannitol (MAN); N-acetyl-glucosamine (NAG); maltose (MAL); gluconate (GNT); caprate (CAP); adipate (ADI); malate (MLT); citrate (CIT); phenyl-acetate (PAC); the API ZYM system tests for the activities of 18 constitutive enzymes: alkaline phosphatase (PHAL); esterase—C4 (EST4); esterase lipase—C8 (EST8); lipase—C 14 (LIP14); leucine arylamidase (LEU); valine arylamidase (VAL); cystine arylamidase (CYS); trypsin (TRY); chymotrypsin (CHY); acid phosphatase (PHAC); α-galactosidase (AGAL); β-galactosidase (BGAL); β-glucuronidase (BGLUR), α-glucosidase (AGLUC); β-glucosidase (BGLUC); N-acetyl-α-glucosaminidase (NACET); α-mannosidase (AMANN); α-fucosidase (AFUC).
A binomial test compared the frequency of positive responses among the pure cultures from each transect (McDonald 2009).
Cultivated bacteria were provisionally identified through the nucleotide sequence of a fragment of their 16S rRNA gene. Genomic DNA was extracted from a colony of each pure culture; the colony was suspended in 50 μl TE buffer (10 mM Tris–HCl, 1 mM EDTA) with 0.6 % Tween 20 and incubated in 80 °C for 20 min. Upon cooling, the suspension was emended with 0.6 μg of proteinase K/ml and incubated at 42 °C for 3 h. Genomic DNA was purified with a Heliconius A&A Biotechnology DNA Clean-up kit. Amplification was performed by PCR (Zdanowski et al. 2004); two sets of primers were used (U515F 5′-GTGCCAGCMGCCGCGGTAA-3′ and 1492R 5′-GGTTACCTTGTTACGACTT-3′; L63F 5′-CAGGCCTAACACATGCAAGTC-3′; and P1387R 5′-GGGCGGTGTGTACAAGGC-3′) (Wang and Qian 2009; Żmuda-Baranowska 2010). Nucleotide sequences were compared by BLAST with those in the NCBI database (Johnson et al. 2008) and using the RDP classifier online program (Cole et al. 2009). Sequences were deposited in GenBank under accession numbers JF728899–JF728980.
Statistics
Simple regression analysis between abundance of the three bacterial fractions (TC, CBcop, and CBoligo) and environmental factors was carried out in STATISTICA v. 9 (StatSoft). Multiple stepwise regression analysis was conducted for three main microbiological parameters (dependent variables): TC, CBcop, and CBoligo. Models were built for two systems: transect 1, transect 2. Analyses were carried out in STATISTICA version 9, (StatSoft).
A canonical correspondence analysis was conducted using a statistical package for Windows v. Canoco 4.5 (ter Braak and Šmilauer 2002). Microbiological data for each sample were analyzed in relation to the environmental background—‘all other data’. To confirm taxonomic diversity in both transects, a DGGE analysis of 10 samples was performed. This allowed use of canonical correspondence analysis to determine differences, but also the impact of the factors determining diversity.
Results
Environmental factors and sampling sites characteristics
Sampling sites were situated along two transects extending 0 m (‘0’) to 672 m (‘E2’) from the edge of Ecology Glacier (Table 1). Soil from TR1 was silty, with sand and fine gravel. Soils from areas with vegetation along TR2 were mixed with plant debris. Ice from the surface of Ecology Glacier was a third sample.
The content of stones and gravel accounted for 6.7–60.3 % (mean 18.4 ± 3.9), sand from 35.2 to 68.7 % (mean 63.2 ± 2.6); dust + silt from 4.5 to 25.6 % (mean 18.6 ± 1.8). The structural components of the terrain between the zones differed slightly along TR1. Terrain furthest from the glacier along TR2 was visually distinguishable.
Total microbial abundance and diversity
Along TR1, with little vegetation, highest TC values were recorded along the edge of the glacier at sampling points 0 (29.48 ± 5.77 × 107) and A1 (39.78 ± 2.52 × 107), with the lowest at point B1 (3.15 ± 0.00 × 107) (Table 2). Along TR2, characterized by richer vegetation, TC values were significantly lower (lowest at A2, 0.41 ± 0.14 × 107), yet they increased with distance from the glacier, reaching a maximum at sampling point F2 (59.06 ± 1.15 × 107).
Highest values of bacterial biomass (Cbact) in TR2 were at a point overgrown with vegetation, while along TR1 the highest values were recorded in the soil at the leading edge of the glacier (Table 2). A modified Shannon index calculated on the basis of cell morphotypes determined by light microscopy ranged from 0.610 to 1.105 along TR2 and correlated with changes in TC (Spearman’s rank correlation, p < 0.05). No such correlation was observed in TR1.
Photoautotrophic microorganisms, including numerically dominant cyanobacteria, diatoms, and unidentified objects of <50 μm in diameter exhibiting red fluorescence, were detected throughout both transects as the smallest TC fraction, with the TAC to TC ratio ranging from 0.05 % at A1 to 31.8 % at D2. Along transects, both the total number of photoautotrophs and cyanobacteria increased from very low values in the youngest Zone I and reached a maximum in the oldest Zone III (Table 2). Note that no cyanobacteria were found at point A1 (TR1). In sample ‘0’, taken at the edge of the glacier front, relatively high numbers of mainly coccoid cells ranging in size from 0.5 to 4.95 μm (probably cyanobacteria) were detected. Filamentous forms dominated in the third zone, in contrast to the coccoid forms prevailing in Zone I. High numbers of diatoms were observed in the third zone, and many of them were covered by bacteria.
Denaturating gradient gel electrophoresis
Molecular analysis by DGGE showed significant differences in taxonomic structure of the soils (Fig. 1). Collectively, 76 OTUs were defined. From the soil samples collected along transects I and II, 40, and 64 OTUs were defined, respectively. In TR1, OTU numbers per sampling site were as follows: ‘0’-17, A1-28, B1-27, C1-19, D1-16, and E1-4. Along TR2, OTU numbers were as follows: A2-10, B2-20, C2-11, D2-30, E2-25, F2-43.
The microbial structure of the soil, soil age, and degree of vegetation cover correlated significantly (Fig. 2). In TR1, the greatest abundance (TC) and taxonomic uniqueness (number of OTUs) were detected in the youngest soil formations, located directly in the glacier foreland. The lowest diversity was determined in the neoglacial Zone III. Within soils in the area of TR2, the highest taxonomic diversity occurred in the oldest soils (Zone III), characterized by the highest vegetation coverage, while the lowest was determined in an area exposed just a few years ago (Zone I).
Culturable bacteria (CB) abundance, diversity, and distribution
Along TR1, CB counts followed the same pattern of TC, with highest numbers in freshly exposed soil (‘0’) and lowest at point B1. In TR2, highest CFU counts were recorded at the oldest exposed sites (E2), and the lowest at points A2 and B2 (Table 2).
Most colonies developed in the later stage of incubation (between 7 and 30 days) and were defined as ‘slow-growers’, as opposed to the minority of CB that grew in the early stages of incubation (up to 7 days), referred to as ‘fast-growers’. The proportion of ‘fast-’ to ‘slow-growers’ was relatively high along TR1’s youngest Zone I (Table 3). In contrast, the highest such proportions in TR2 occurred in the area farthest from the glacier, in places covered with vegetation.
Significant differences occurred among the morphophysiological characteristics of bacteria isolated along the two transects (Fig. 3). With the exception of twelve characteristics, most features were significantly more frequent in bacteria cultivated from TR1. Psychrophiles (those whose growth was inhibited above 20 °C) occurred along both transects, but were clearly more prevalent in TR2 (80 %). Note that psychrophiles accounted for only 60 % of the total number of strains isolated from the glacier surface. Almost 40 % of bacteria cultures from TR1 could reduce NO3 − to NO2 −, while less than 10 % of such strains from TR2 could do so. Up to 30 % of bacteria cultures prepared from TR1-expressed cytochrome oxidase activity (OX+). A high percentage of oxidase positive strains were isolated from the sample in direct contact with the glacier at that time.
Differences in the incidence of morphophysiological positive tests among the cultivated bacteria isolated from two transects: NS not significant, TR transect, N number of isolates calculated on the basis of the counts of similar colonies of which representative was tested. Significance level (*p < 0.05, **p < 0.01, ***p < 0.001) between frequency of positive responses to tests shown by bacterial communities. Tests conducted: ability to grow at 4, 22, 32 °C; pigmented colony counts (PIGM), Gram stain (GRAM), cell morphology (ROD); reduction of nitrate to nitrite (NO3), and to nitrogen (N2); indole production (TRP); fermentation of glucose (GLU_F); arginine dihydrolase (ADH); urease ((URE); β-glucosidase (ESC); gelatin hydrolysis (GEL); cytochrome oxidase (OX); assimilation tests: glucose (GLU); arabinose (ARA), mannose (MNE); mannitol (MAN); N-acetyl-glucosamine (NAG); maltose (MAL); gluconate (GNT); caprate (CAP); adipate (ADI); malate (MLT); citrate (CIT); phenyl-acetate (PAC); enzyme activity tests: alkaline phosphatase (PHAL); esterase—C4 (EST4); esterase lipase—C8 (EST8); lipase—C 14 (LIP14); leucine arylamidase (LEU); valine arylamidase (VAL); cystine arylamidase (CYS); trypsin (TRY); chymotrypsin (CHY); acid phosphatase (PHAC); α-galactosidase (AGAL); β-galactosidase (BGAL); β-glucuronidase (BGLUR), α-glucosidase (AGLUC); β-glucosidase (BGLUC); N-acetyl-α-glucosaminidase (NACET); α-mannosidase (AMANN); α-fucosidase (AFUC)
The number of cultivated isolates that could assimilate single carbon sources varied between transects 1 and 2. The mean number of positive responses in assimilation of 12 different carbohydrates and organic acids, meaning those that could use these as sole sources, was statistically higher among bacteria cultivated from TR1. Relatively few of the strains isolated from the surface of the glacier assimilated these carbohydrates. The percentage of cultivated strains expressing glycoside hydrolase activities was highest among bacteria cultivated from TR1. Strains from Zones II and III were most likely to express glycosidases. A low percentage of strains possessing glycosidases was observed on the surface and in contact with the glacier. TR1 hosts bacterial communities characterized by a broader range in constitutive enzyme activity and sole carbon source metabolism, unlike that from stable TR2. Only 11 of the 46 traits occurred with equal frequency in both groups. These were the rarely expressed traits.
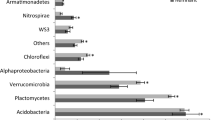
The 82 pure cultures whose 16S rRNA genes were partially sequenced fell into 56 OTUs at the 98 % sequence identity level (Table 4). Seventy-five cultures were classified to the genus level. Actinobacteria were the most numerous and also the most diverse (41 strains, 26 OTUs). Less numerous and less diverse were Bacteroidetes (11 strains, 9 OTUs), Alphaproteobacteria (12 strains, 6 OTUs), Betaproteobacteria (8 strains, 6 OTUs), Firmicutes (6 strains, 3 OTUs), and Gammaproteobacteria (4 strains, 4 OTUs). The nearest neighbors in BLAST searches were from cold environments, such as Antarctica, the Arctic, Himalayas, Alps, and deep-sea sediments.
The dominant phylum among bacteria grown from TR1 and the surface of the glacier was the Actinobacteria (Table 4) represented by Frigoribacterium spp.—found on the glacier, Leifsonia—present mainly at the interface between the glacier and Arthrobacter—mostly isolated from Zones II and III of the first transect, but also Zone III of the second transect.
Proteobacteria were most abundant among bacteria cultivated from TR2. The Alphaproteobacteria (Sphingomonas, Brevundimonas) occurred in large quantities at sites that became ice-free in the last 10 years, but their numbers decreased with distance from the glacier. The Betaproteobacteria represented by Polaromonas were detected on the glacier and along TR2. Herminiimonas (Betaproteobacteria) numerically dominated at sites F2 and E2—the earliest exposed as the glacier retreated. Pseudomonas (Gammaproteobacteria) was also present at the latter site. Bacteroidetes (Flavobacterium, Pedobacter) were present in moderate quantities in sites in early and middle stages of glacier foreland succession. Firmicutes (Sporosarcina) were found in both transects but only in areas far from the glacier.
Statistics
Simple regression analysis between abundance of the three bacterial fractions (TC, CBcop, and CBoligo) and environmental factors showed that in TR1, these significantly correlated with the soil pH (Table 5). Water and organic carbon content correlated positively with TC only in TR2, whereas the mineral content (P2O5, K, Mg, Ca) in TR1. Time since exposure negatively correlated with TC in TR1 and positively with CB in TR2. Relatively large soil particles (stones, sand) and plant cover had an overall positive effect on the microbiocenosis.
Multiple regression analysis identified those independent variables whose influence was most significant (Table 6). In TR1, Ca and P2O5 contents were significant for TC, and Na content for the CB. In TR2, it was the water content and soil age (time since exposure) that had a major impact on the microbiocenosis.
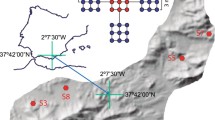
The statistical analysis of principal components (CCA) showed a significant correlation between the taxonomic structure (of OTUs) and selected biological and physicochemical parameters of soils (Fig. 4). The main factors determining the microbial structure of the soil in TR1 were physicochemical properties, especially pH. In soils covered with vegetation (TR2), significant factors affecting the taxonomic structure were mainly biological, particularly the abundance of photoautotrophs (TAC).
Canonical correspondence analysis of denaturating gradient gel electrophoresis bands and environmental data. Empty square transect 1, solid square transect 2, solid circle control, bacterial total count (TC), total photoautotrophic count (TAC), cyanobacteria count (CYAN), cultivated copiotrophic bacteria count (CBcop), cultivated oligotrophic bacteria count (CBoligo), total bacterial carbon (TCB), loss on ignition (OM)
Discussion
Bacterial communities in postglacial soils of Ecology Glacier (King George Island, Antarctica) were investigated in terms of abundance, physiology, and taxonomic affiliation. Our findings provide a description of changes in the glacier’s edge, as well as a comparison of soil microbiocenoses in two transects with different ground stability and plant coverage along lateral moraines.
Glacier ice–foreland interactions
Biogeochemical data were in line with the literature reports on postglacial areas (Bardgett et al. 2007; Nemergut et al. 2007). However, total organic carbon was lower than in the high Arctic (Schmidt et al. 2008). The high total carbon contents at point ‘0’ (close to the glacier) were striking, considering the lower levels 100–200 m away from the glacier, where microbial photosynthesis would be expected to provide organic carbon. This might be attributable to the enriching influence of supraglacial meltwater (such as from cryoconite holes) or ancient subglacial sediment carbon (Stibal et al. 2006; Bardgett et al. 2007). The highest TC and CFU counts were determined in a zone closest to the glacier (point ‘0’). The CB/TC ratio was also high here and might be supported by enrichment of the soil by supraglacier meltwater bacteria (Christner et al. 2003; Mueller and Pollard 2004; Grzesiak et al. 2009). Supraglacial meltwater input may also explain the relatively high count of cyanobacteria at point ‘0’, while their numbers in the newly exposed glacial terrain of Zone I and II, farther from the glacier, showed low values. This is in line with Tscherko et al. (2003a) who reported that heterotrophic microbial communities occur in newly exposed glacial terrain before autotrophic communities. Initial taxonomic structure comparison analysis of soil from the Ecology Glacier foreland and from the glacier’s surface ice by DGGE showed they share 30 % of OTUs (by taxonomic affiliation) with Zone I. However, this number decreases markedly with distance from the glacier, with 6 and 4 % of shared OTUs for Zones II and III, respectively. This indicates that glacial runoff derived bacteria play a role only in early stages of postglacial succession.
Ribosomal DNA analysis of bacteria cultivated from soil close to the glacier, and from the glacial ice, did not detect any common genera between the two. This might reflect the very different pH values of the glacial ice and freshly exposed forefield soil, or high Na+ content of the soil. The high TC and composition of the culturable bacteria community in this soil were similar even 2 years after deglaciation (sampling point A1). Therefore, high culturable bacteria counts might be derived from below the glacier. For example, culturable bacteria counts in basal sediment beneath a high Arctic glacier may reach 107 ml−1 (Skidmore et al. 2000; Cheng and Foght 2007). Supraglacial melt water enrichment thus appears more nutritional than microbial in nature. This explanation is supported by the high numbers of copiotrophic and fast-growing bacteria (r strategy) at the glacier edge. A similar pattern of development per day of CB in the Ecology Glacier forefield has been reported before (Zdanowski and Węgleński 2001). These observations are consistent with the hypothesis of Sigler and Zeyer (2004), that bacteria able to quickly respond to disturbance and tolerate stress are more abundant in younger soils, where conditions favor r-strategists, typical of opportunistic bacteria (Sigler et al. 2002). It should be noted that we incubated samples for cultivation of bacteria under conditions that differed from those used by Sigler et al. (2002), since we avoided incubation temperatures that would be inappropriate for most psychrophilic and psychrotrophic microbes, that is, we used 4 °C instead of 25 °C.
Microbial community along transect 2
A classic succession pattern in terms of soil elements, microbial abundance, and bacterial taxonomic richness was evident along transect 2 (Schipper et al. 2001; Sigler et al. 2002; Schmidt et al. 2008). In this nutrient-limited postglacial environment, the contents of biogenic elements gradually increased. Total and photoautotrophic cell counts followed this trend, reaching a maximum in well-formed soils in Zone III. Sattin et al. (2009) describe a similar observation based on clone libraries during their work on Mendenhall Glacier foreland soils (USA). Counts at sampling points E2 and F2 could have been influenced by significant numbers of edaphic ciliates which control bacterial populations (Mieczan, unpublished).
Stable ground conditions along TR2, with minor freeze–thaw events, have enabled development of plant coverage, which according to the CCA analysis positively influenced bacterial communities along this transect (Fig. 4). Community richness determined by DGGE analysis increased with distance from the glacier (Fig. 2); a phenomenon recognized in ecological studies where the number of species in an undisturbed environment tends to increase with age (Walker and del Moral 2003). The presence of plants and microbial photoautotrophs is especially beneficial for the culturable fraction of the community as indicated by the relatively high CB to TC ratio. This is also reflected in the phenotypic properties of strains isolated from TR2. Plant coverage on TR2 promoting more stable and lower temperatures would explain significantly higher numbers of psychrophilic bacteria along this transect. Moreover, bacteria cultivated from TR2 expressed a narrow range of constitutive enzymes and utilized only a few carbon sources (Fig. 3), implying a stable and perhaps single source of nutrients. Cultivated bacteria from transect 2, that was less affected by cryoturbation than transect 1, were taxonomically more diverse in terms of 16Sr RNA gene nucleotide sequences. That no genus dominated the chronosequence indicates favorable conditions for bacterial colonization, and greater likelihood of successful allochthonous introductions.
Members of the generally oligotrophic genus Brevundimonas were cultivated from Zones I and II in both transects, but reached very high numbers only in transect 2. Brevundimonas, although detected in low numbers at the glacier’s edge, developed large populations later in the succession process. Members of this genus have been isolated from various polar habitats, for example, glacial snow in summer (Amato et al. 2006), Antarctic soil and lakes, Arctic deep ice core and snow cover, subglacial outflow waters (Cheng and Foght 2007), and also from the Antarctic aerosol over King George Island (Miteva et al. 2004; Amato et al. 2006; González-Toril et al. 2009). Polaromonas sp. seems to be the only surface ice derived genus among the CB bacteria in the moraine soil older than 30 years. However, it was only cultivated here from transect 2, so cryoturburbation and large temperature shifts in TR1 may not be favorable for members of this genus.
A non-opportunistic growth strategy and dependence on plant-specific growth factors may be the reason for the high percentage of strains lost during recultivation from late succession period sites (C2, D2, E2, F2). Those samples did, however, yield bacteria from genera considered plant-associated and/or plant-dependent, such as Sporosarcina, Bacillus, Pedobacter, Herminiimonas, Sphingomonas, and Rhodococcus, psychrophilic members of which have been isolated from the lichen and moss rhizospheres in polar regions (Männisto and Häggblom 2006; Lang et al. 2007; Selbmann et al. 2010). Furthermore, Herminiimonas sp. has been reported to assimilate oxalic acid produced by lithophytic lichens (Lang et al. 2007). Thus, Herminiimonas spp. at the humus-rich F2 sampling point may be associated with lichen communities and their production of secondary metabolites. Sphingomonas spp. (Alphaproteobacteria) also are often found in association with plants (in the rhizosphere) (White et al. 1996); some are nitrogen fixers, suggesting a potential role as plant symbionts (Doty et al. 2009). Pedobacter spp. have been isolated from soils, permafrost, glacial, and near glacial environments and from decaying lichens (Bai et al. 2006; Männisto and Häggblom 2006; Hansen et al. 2007; Yoon et al. 2007; Simon et al. 2009). Members of the Sphingobacteriaceae such as Pedobacter sp. can utilize carbohydrates that are plant derived (Liebner et al. 2008).
Microbial community along transect 1
Transect 1 was selected because of its lack of vegetation, on the premise that this could influence microbial communities. However, it was quickly recognized as undergoing severe cryoturbation, and a process that in many ways impairs the postglacial soil development and bacterial succession determined along TR2. Cryoturbation consequences for soil and microbial community development are clearly visible. The main factors affecting soil microbiocenoses are most likely the freeze–thaw cycle and lack of plant cover. The latter results inter alia in a greater daily temperature range. The CCA analysis, however, showed that pH in these circumstances is of crucial importance to the bacterial community. In this respect, a lack of rock-weathering organic acids produced by some mosses and lichens would explain why the pH of the soils lacking vegetation along TR1 never dropped below 8.
Despite cryoturbation and the lack of plant cover, TC and CB values were relatively high in all TR1 soil samples. Counts of edaphic ciliates as low as four individuals per gram of soil would not be expected to significantly affect the size of the bacterial communities (Mieczan, unpublished). Taxonomic diversity among the bacterial community diminished with distance from the glacier (Fig. 2). Pure cultures of bacteria isolated from TR1 expressed a wide range of enzymes and assimilated various carbon compounds, useful characteristics for life in an environment where biogenic elements show a ‘patchy’ distribution.
Members of the Leifsonia genus numerically dominated the CB community in the very early stages of succession in transect 1. This is rather surprising considering their copiotrophic and photoautotroph-associated nature (Evtushenko and Takeuchi 2006). It is more likely that the isolates cultivated here were of subglacial origin, as no representatives of the genus were found in surface ice or anywhere else along the transect. For example, recently described Leifsonia species were isolated from cold habitats such as soil near a Himalayan Glacier, Southern Ocean sediment (Pindi et al. 2009), Antarctic pond cyanobacterial mat (Reddy et al. 2003), and Spitsbergen permafrost (Hansen et al. 2007), suggesting this genus is well adapted to life in polar and alpine ecosystems. One of its ecological functions could be supplying autotrophs with phosphorus and metal ions of biological importance such as Fe, Mn, Ca, K, and Mg by weathering of poorly available sources (insoluble mineral deposits). In this respect, the bioleaching properties of Leifsonia spp. have been widely recognized (Hansen et al. 2007; Delvasto et al. 2008; Frey et al. 2010).
A sharp drop in total bacterial abundance and change in the taxonomic composition of the bacteria cultivated occurred between sampling points A1 and B1, with a shift from Leifsonia to Arthrobacter. We posit that this may be a response to an environmental factor such as soil mineral composition change (causing a pH drop), indicating a limit of glacial influence. Due to their extreme resistance to drying, starvation, temperature shifts, pH changes, and nutritional versatility (Jones and Keddie 2006; Mongodin et al. 2006), Arthrobacter strains replace Leifsonia spp. in such conditions and are clearly recognized as pioneer bacteria in this study. Along TR1, for example, these bacteria comprised up to 98 % of the CB counts, testimony to their durability, and survival under cryoturbation conditions that would be lethal to other secondary succession bacteria. However, a succession pattern exists even in this genus (Table 4). A penguin rookery close to Ecology Glacier (Trivelpiece et al. 1990; Ciaputa and Sierakowski 1999) is a likely source of Arthrobacter spp. Studies have already shown Arthrobacter species engaged in guano decomposition are abundant, nutritionally versatile, and adaptable (Zdanowski et al. 2005; Żmuda-Baranowska 2010).
Summary
Our findings indicate that recently deglaciated soils host abundant microorganisms and biogenic elements which appear to be the result of sub- and supraglacial influences. This unique environment maintains its character even several years after deglaciation. However, the microbiological contribution to later succession stages seems limited. A more intense allochthonous colonization begins when glacier-associated factors no longer determine microbial community composition, and ‘pioneer-microorganisms’ start to flourish. Diversity and properties of postglacial microbiocenoses result not only from soil age and composition, but also from differences in soil stability. An undisturbed ground enables vegetation growth and promotes a more taxonomically diverse microbial community, with types of bacteria being plant-associated and less resistant to temperature fluctuations. Cryoturbation, which hinders colonization by plants, causes a gradual biodiversity impoverishment and emergence of dominance by just a few types of bacteria which are nutritionally versatile and very resistant to abiotic conditions. Therefore, we recognize cryoturbation as an extremely important environmental factor shaping microbial community structure and development in postglacial soils.
References
Amato P, Hennebelle R, Magand O, Sancelme M, Delort AM, Barbante C, Bourton C, Ferrari C (2006) Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol Ecol 59:255–264
Bai Y, Yang D, Wang J, Xu S, Wang X, An L (2006) Phylogenetic diversity of culturable bacteria from alpine permafrost in the Tianshan Mountains, northwestern China. Res Microbiol 157:741–751
Bardgett RD, Richter A, Bol R, Garnett MH, Bäumler R, Xingliang X, Lopez-Capel E, Manning DAC, Hobbs PJ, Hartley IR, Wanek W (2007) Heterotrophic microbial communities use ancient carbon following glacial retreat. Biol Lett 3:487–490
Beyer L, Bockheim J, Campbell IB, Claridge GGC (1999) Properties, genesis and global significance of Antarctic cryosols. Antarct Sci 11:387–398
Beyer L, Blume HP, Bölter M, Kappen L, Seppelt RD (2002) Soil ecology in relation to plant patterns. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 375–388
Birkenmajer K (2002) Retreat of Ecology Glacier, Admiralty Bay, King George Island (South Shetland Island, West Antarctica) 1956–2001. Bull Pol Acad Sci Earth Sci 50:5–19
Blume HP, Kuhn D, Bölter M (2002a) Soils and soilscapes. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 91–112
Blume HP, Beyer L, Kalk E, Kuhn D (2002b) Weathering and soil formation. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 115–137
Bölter M, Blume HP (2002) Soils as habitats for microorganisms. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 285–299
Cheng SM, Foght JM (2007) Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol Ecol 59:318–330
Christner BC, Kvitko BH, Reeve JN (2003) Molecular identification of bacteria and eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7:177–183
Ciaputa P, Sierakowski K (1999) Long-term population changes of Adélie, chinstrap and gentoo penguins in the regions of SSSI No.8 and SSSI No.34, King George Island, Antarctica. Pol Polar Res 20:55–365
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/nar/gkn879
Crump BC, Hobbie JE (2005) Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr 50:1718–1729
Crump BC, Kling GW, Bahr M, Hobbie JE (2003) Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol 69:2253–2268
Crump BC, Peranteau C, Beckingham B, Cornwell JC (2007) Respiratory succession and community succession of bacterioplankton in seasonally anoxic estuarine waters. Appl Environ Microbiol 73:6802–6810
Darmody RG, Allen CE, Thorn CE (2005) Soil topochronosequences at Storbreen, Jotunheimen, Norway. Soil Sci Soc Am J 69:1275–1287
Delvasto P, Valverde A, Ballester A, Munoz JA, González F, Blázquez ML, Igual JM, García-Balboa C (2008) Diversity and activity of phosphate bioleaching bacteria from a high-phosphorus iron ore. Hydrometallurgy 92:124–129
Doty SL, Oakley B, Xin G, Kang JW, Singleton GL, Khan Z, Vajzovic A, Staley JT (2009) Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47:23–33
Dowgiałło A (1975) Chemical composition of an animal body and its food. In: Grodziński W, Klekowski RZ, Duncan A (eds) Methods for ecological bioenergetics. IBP handbook. Blackwell, Oxford, pp 160–199
Evtushenko LI, Takeuchi M (2006) The family Microbacteriaceae. In: Dvorkin M, Falkov S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes, vol 3. Springer, New York, pp 1020–1098
Fenchel T, Hemmingsen BB (1974) Manual of microbial ecology. Akademisk Forlag/University of Aarhus, Copenhagen/Aarhus, p 208
Frey B, Rieder SR, Brunner I, Plötze M, Koetzsch B, Lapanje A, Brandl H, Furrer G (2010) Weathering-associated bacteria from the Damma Glacier forefield: physiological capabilities and impact on granite dissolution. Appl Environ Microbiol 78:4788–4796
González-Toril E, Amils R, Delmas RJ, Petit J-R, Komárek J, Elster J (2009) Bacterial diversity of autotrophic enriched cultures from remote, glacial Antarctic, Alpine and Andean aerosol, snow and soil samples. Biogeosciences 6:33–44
Grzesiak J, Żmuda-Baranowska MJ, Borsuk P, Zdanowski MK (2009) Microbial community at the front of Ecology Glacier (King George Island, Antarctica): Initial observations. Pol Polar Res 30:37–47
Gurienowich T (1995) Osobiennosti razvitia bakterioplanktona i bakteriobentosa raznotipnych rybohoziastviennych vodojemov. (Characteristic and development of bakterioplankton and benthic bacterial communities in different types of fish pond). PhD thesis, The State Scientific Institute for Fisheries, Sankt-Petersburg, Russia (in Russian)
Hansen AA, Herbert RA, Mikkelsen K, Jensen LL, Kristoffersen T, Tiedje JM, Lomstein BA, Finster KW (2007) Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, northern Norway. Environ Microbiol 9:2870–2884
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi:10.1093/nar/gkn201
Jones D, Keddie RM (2006) The genus Arthrobacter. In: Dvorkin M, Falkov S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes, vol 3. Springer, New York, pp 945–960
Klement Z, Rudolph K, Sands DC (1990) Methods in phytobacteriology. Akademiai Kiado, Budapest, p 568
Lang E, Swiderski J, Stackebrandt E, Schumann P, Sproer C, Sahin N (2007) Herminiimonas saxobsidens sp. nov., isolated from a lichen-colonized rock. Int J Syst Evol Microbiol 57:2618–2622
Liebner S, Harder J, Wagner D (2008) Bacterial diversity and community structure in polygonal tundra soils from Samoylov Island, Lena Delta, Siberia. Int Microbiol 11:195–202
Männisto MK, Häggblom MM (2006) Characterization of psychrotolerant heterotrophic bacteria from Finnish Lapland. Syst Appl Microbiol 29:229–243
McDonald JH (2009) Handbook of biological statistics, 2nd edn. Sparky House Publishing, Baltimore, pp 24–32
Miteva VI, Sheridan P, Brenchley J (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70:202–213
Mongodin EF, Shapir N, Daugherty SC, DeBoy RT, Emerson JB, Shvartzbeyn A, Radune D, Vamathevan J, Riggs F, Grinberg V, Khouri H, Wackett LP, Nelson KE, Sadowsky MJ (2006) Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet 12:2094–2106
Mueller DR, Pollard WH (2004) Gradient analysis of glacial cryoconite ecosystems: a bipolar comparison. Polar Biol 27:66–74
Nemergut DR, Anderson SP, Cleveland CC, Martin AP, Miller AE, Seimon A, Schmidt SK (2007) Microbial community succession in an unvegetated, recently-deglaciated soil. Microb Ecol 53:110–122
Nübel U, Garcia-Pichel F, Kühl M, Muyzer G (1999) Quantifying microbial diversity: morphotypes, 16SrRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol 65:422–430
Ogram A, Feng X (1997) Methods of soil microbial community analysis. In: Hurst CJ, Knudsen GH, McInerney MJ, Stetzenbach LD, Walter MV (eds) Manual of environmental microbiology. ASM Press, Washington, pp 422–430
Olech MA (2002) Plant communities on King George Island. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 215–231
Orchard MA, Corderoy DM (1983) Influence of environmental factors on the decomposition of penguin guano in Antarctica. Polar Biol 1:199–204
Parnikoza IYu, Inozemtseva DM, Tyshenko OV, Mustafa O, Kozeretska IA (2008) Antarctic herb tundra colonization zones in the context of ecological gradient of glacial retreat. Ukr Botany Journ 4:504–512
Pindi PK, Kishore KH, Reddy GS, Shivaji S (2009) Description of Leifsonia kafniensis sp. nov. and Leifsonia antarctica sp. nov. Int J Syst Evol Microbiol 59:1348–1352
Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943–948
Pudełko R (2003) Topographic map of the SSSI No. 8, King George Island, West Antarctica. Pol Polar Res 23:53–60
Pudełko R (2008) Two new topographic maps for sites of scientific interest on King George Island, West Antarctica. Pol Polar Res 29:291–297
Putland JN, Rivkin RB (1999) Influence of storage mode and duration on the microscopic enumeration of Synechococcus from cold coastal environment. Aquat Microb Ecol 17:191–199
Rassoulzadegan F, Sheldon RW (1986) Predator–prey interactions of nanozooplankton and bacteria in an oligotrophic marine environment. Limnol Oceanogr 31:1010–1021
Reddy GSN, Matsumoto GI, Shivaji S (2003) Sporosarcina macmurdoensis sp. nov., from a cyanobacterial mat sample from a pond in the McMurdo Dry Valleys, Antarctica. Int J Syst Evol Microbiol 53:1363–1367
Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, Robeson MS, Ascarrunz N, Nemergut DR (2009) Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol 47:673–681
Schipper LA, Degens BP, Sparling GP, Duncan LC (2001) Changes in microbial heterotrophic diversity along five plant successional sequences. Soil Biol Biochem 33:2093–2103
Schmalenberger A, Noll M (2010) Shifts in desulfonating bacterial communities along a soil chronosequence in the forefield of a receding Glacier. FEMS Microbiol Ecol 71:208–217
Schmidt SK, Reed SC, Nemergut DR, Grandy AS, Cleveland CC, Weintraub MN, Hill AW, Costello EK, Meyer AF, Neff JC, Martin AM (2008) The earliest stages of ecosystem succession in high-elevation (5000 meters above sea level), recently deglaciated soils. Proc R Soc B 275:2793–2802
Selbmann L, Zucconi L, Ruisi S, Grube M, Cardinale M, Onofri S (2010) Culturable bacteria associated with Antarctic lichens: affiliation and psychrotolerance. Polar Biol 33:71–83
Seppelt RD (2002) Wilkes Land (Casey Station). In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological studies, vol 154. Springer, Berlin, pp 41–46
Sieracki ME, Johnson PW, JMcN Sieburth (1985) Detection, enumeration, and sizing of planktonic bacteria by image-analyzed epifluorescence microscopy. Appl Environ Microbiol 49:799–810
Sigler WV, Zeyer J (2004) Colony-forming analysis of bacterial community succession in deglaciated soils indicates pioneer stress-tolerant opportunists. Microb Ecol 48:316–323
Sigler WV, Crivii S, Zeyer J (2002) Bacterial succession in glacial forefield soils characterized by community structure, activity and opportunistic growth dynamics. Microb Ecol 44:306–316
Simon M, Azam F (1989) Protein content and protein synthesis rates of planktonic bacteria. Mar Ecol Prog Ser 51:201–213
Simon C, Herath J, Rockstroh S, Daniel R (2009) Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl Environ Microbiol 75:2964–2968
Skidmore ML, Foght JM, Sharp MJ (2000) Microbial life beneath a high Arctic glacier. Appl Environ Microb 66:3214–3220
Smykla J, Wołek J, Barcikowski A (2007) Zonation of vegetation related to penguin rookeries on King George Island, Maritime Antarctic. Arct Antarct Alp Res 39:143–151
Stibal M, Šabacká M, Kaštovská K (2006) Microbial communities on glacier surfaces in Svalbard: impact of physical and chemical properties on abundance and structure of cyanobacteria and algae. Microb Ecol 52:644–654
Świątecki A (1997) Zastosowanie wskaźników bakteriologicznych w ocenie wód powierzchniowych. (Application of bacteriological indicators in surficial water quality assessment). WSP Olsztyn (in Polish), pp 1–105
Tatur A, Myrcha A, Niegodzisz J (1997) Formation of abandoned penguin rookery ecosystems in the maritime Antarctic. Polar Biol 17:405–417
ter Braak CJF, Šmilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca
Tibbles BJ, Harris JM (1996) Use of radiolabelled thymidine and leucine to estimate bacterial production in soils from continental Antarctica. Appl Environ Microb 62:694–701
Tockner K, Malard F, Uehlinger U, Ward JV (2002) Nutrients and organic matter in a glacial river–floodplain system (Val Roseg, Switzerland). Limnol Oceanogr 47:266–277
Trivelpiece WZ, Trivelpiece SG, Geupel GR, Kjelmyr J, Volkman NJ (1990) Adélie and chinstrap penguins: their potential as monitors of Southern Ocean marine ecosystems. In: Kerry KR, Hempel G (eds) Antarctic ecosystems, ecological change and conservation. Springer, Berlin/Heidelberg, pp 191–202
Tscherko D, Rustemeier J, Richter A, Wanek W, Kandeler E (2003a) Functional diversity of the soil microflora in primary succession across two glacier forelands. Eur J Soil Sci 54:685–696
Tscherko D, Bölter M, Beyer L, Chen J, Elster J, Kandeler E, Kuhn D, Blume HP (2003b) Biomass and enzyme activity of two soil transects at King George Island, maritime Antarctica. Arct Antarct Alp Res 35:34–47
Victoria FC, Pereira AB, Costa DP (2009) Composition and distribution of moss formations in the ice-free areas adjoining the Arctowski region, Admiralty Bay, King George Island, Antarctica. Sér Bot 1:81–91
Walker LR, del Moral R (2003) Primary succession and ecosystem rehabilitation. Cambridge University Press, Cambridge, pp 332–333
Wang Y, Qian P-Y (2009) Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PloS ONA 10:1–9
White DC, Sutton SD, Ringelberg DB (1996) The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol 7:301–306
Yoon M-H, Ten LN, Im W-T, Lee S-T (2007) Pedobacter panaciterrae sp. nov., isolated from soil in South Korea. Int J Syst Evol Microbiol 57:381–386
Zdanowski MK, Węgleński P (2001) Ecophysiology of soil bacteria in the vicinity of Henryk Arctowski Station, King George Island, Antarctica. Soil Biol Biochem 33:819–829
Zdanowski MK, Węgleński P, Golik P, Sasin JM, Borsuk P, Żmuda MJ, Stankovic A (2004) Bacterial diversity in Adelie penguin, Pygoscelis adeliae guano: molecular and morphological approaches. FEMS Microbiol Ecol 50:163–173
Zdanowski MK, Żmuda MJ, Zwolska I (2005) Bacterial role in the decomposition of marine-derived material (penguin guano) in the terrestrial maritime Antarctic. Soil Biol Biochem 37:581–595
Żmuda-Baranowska MJ (2010) Diversity of bacteria participating in marine derived organic matter degradation in Antarctic and Arctic shore areas (in Polish). PhD thesis, Department of Antarctic Biology, Polish Academy of Sciences, Warsaw
Acknowledgments
This research was supported by the Polish Ministry of Science and Higher Education (grant IPY/26/2007) and by the National Centre for Science in Krakow PL (grant N N304 106940). We are very grateful to Prof. Stuart Donachie (University of Hawaii) and Prof. Arthur A. Vandenbark (OHSU Multiple Sclerosis Center) for their comments and language editing during preparation of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zdanowski, M.K., Żmuda-Baranowska, M.J., Borsuk, P. et al. Culturable bacteria community development in postglacial soils of Ecology Glacier, King George Island, Antarctica. Polar Biol 36, 511–527 (2013). https://doi.org/10.1007/s00300-012-1278-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-012-1278-0