Abstract
Key message
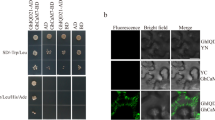
In GhCFE5 homoeologs, GhCFE5D interacted with more actin homologs and stronger interaction activity than GhCFE5A. GhCFE5D - but not GhCFE5A -overexpression severely disrupted actin cytoskeleton organization and significantly suppressed cell elongation.
Abstract
Homoeologous genes are common in polyploid plants; however, their functional divergence is poorly elucidated. Allotetraploid Upland cotton (Gossypium hirsutum, AADD) is the most widely cultivated cotton; accounting for more than 90 % of the world’s cotton production. Here, we characterized GhCFE5A and GhCFE5D homoeologs from G. hirsutum acc TM-1. GhCFE5 homoeologs are expressed preferentially in fiber cells; and a significantly greater accumulation of GhCFE5A mRNA than GhCFE5D mRNA was found in all tested tissues. Overexpression of GhCFE5D but not GhCFE5A seriously inhibits the Arabidopsis hypocotyl and root cell elongation. Yeast two-hybrid assay and bimolecular fluorescence complementation (BiFC) analysis showed that compared with GhCFE5A, GhCFE5D interacts with more actin homologs and has a stronger interaction activity both from Arabidopsis and Upland cotton. Interestingly, subcellular localization showed that GhCFE5 resides on the cortical endoplasmic reticulum (ER) network and is colocalized with actin cables. The interaction activities between GhCFE5 homoeologs and actin differ in their effects on F-actin structure in transgenic Arabidopsis root cells. The F-actin changed direction from vertical to lateral, and the actin cytoskeleton organization was severely disrupted in GhCFE5D-overexpressing root cells. These data support the functional divergence of GhCFE5 homoeologs in the actin cytoskeleton structure and cell elongation, implying an important role for GhCFE5 in the evolution and selection of cotton fiber.







Similar content being viewed by others
Abbreviations
- CaMV:
-
Caulifower mosaic virus
- DAG:
-
Day after germination
- DPA:
-
Day post anthesis
- DUF:
-
Domain of unknown function
- SNPs:
-
Single-nucleotide polymorphisms
- NPT-II:
-
Neomycin phosphotransferase II
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green fluorescent protein
- RFP:
-
Red fluorescent protein
References
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Most M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Vainola R, Wolf JB, Zinner D (2013) Hybridization and speciation. J Evol Biol 26:229–246
Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100:4649–4654
Applequist WL, Cronn R, Wendel JF (2001) Comparative development of fiber in wild and cultivated cotton. Evol Dev 3:3–17
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1148–1152
Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15:57–71
Chen CY, Cheung AY, Wu HM (2003) Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15:237–249
Chen M, Ha M, Lackey E, Wang J, Chen ZJ (2008) RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178:1845–1858
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42:443–461
Endrizzi JE, Turcotte EL, Kohel RJ (1985) Genetics, cytology and evolution of Gossypium. Adv Genet 23:271–375
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Gilliland LU, Kandasamy MK, Pawloski LC, Meagher RB (2002) Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol 130:2199–2209
Gilliland LU, Pawloski LC, Kandasamy MK, Meagher RB (2003) Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J 33:319–328
Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5:3050
Guo W, Cai C, Wang C, Han Z, Song X, Wang K, Niu X, Wang C, Lu K, Shi B, Zhang T (2007) A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176:527–541
Guo W, Cai C, Wang C, Zhao L, Wang L, Zhang T (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genom 9:314
Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA 106:17835–17840
Ha M, Ng DW, Li WH, Chen ZJ (2011) Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res 21:590–598
Han LB, Li YB, Wang HY, Wu XM, Li CL, Luo M, Wu SJ, Kong ZS, Pei Y, Jiao GL, Xia GX (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25:4421–4438
Hu Z, Song N, Xing J, Chen Y, Han Z, Yao Y, Peng H, Ni Z, Sun Q (2013) Overexpression of three TaEXPA1 homoeologous genes with distinct expression divergence in hexaploid wheat exhibit functional retention in Arabidopsis. PLoS One 8:e63667
Hutchinson JB (1951) Intra-specific differentiation in Gossypium hirsutum. Heredity 5:161–193
Jiang JX, Zhang TZ (2003) Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method. Cotton Sci 15:166–167
Jiang C, Wright RJ, El-Zik KM, Paterson AH (1998) Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc Natl Acad Sci USA 95:4419–4424
Kandasamy MK, McKinney EC, Meagher RB (2009) A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21:701–718
Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188:263–272
Lashermes P, Combes MC, Hueber Y, Severac D, Dereeper A (2014) Genome rearrangements derived from homoeologous recombination following allopolyploidy speciation in coffee. Plant J 78:674–685
Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Ann Bot 100:1391–1401
Leitch AR, Leitch IJ (2008) Genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Li S, Blanchoin L, Yang Z, Lord EM (2003) The putative Arabidopsis arp2/3 complex controls leaf cell morphogenesis. Plant Physiol 132:2034–2044
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Li Y, Jiang J, Li L, Wang XL, Wang NN, Li DD, Li XB (2013) A cotton LIM domain-containing protein (GhWLIM5) is involved in bundling actin filaments. Plant Physiol Biochem 66:34–40
Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, Chen W, Liang X, Shang H, Liu W, Shi C, Xiao G, Gou C, Ye W, Xu X, Zhang X, Wei H, Li Z, Zhang G, Wang J, Liu K, Kohel RJ, Percy RG, Yu JZ, Zhu YX, Wang J, Yu S (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Li L, Li Y, Wang NN, Li Y, Lu R, Li XB (2015) Cotton LIM domain-containing protein GhPLIM1 is specifically expressed in anthers and participates in modulating F-actin. Plant Biol (Stuttg) 17:528–534
Liu T, Tian J, Wang G, Yu Y, Wang C, Ma Y, Zhang X, Xia G, Liu B, Kong Z (2014) Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr Biol 24:2708–2713
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Lv F, Wang H, Wang X, Han L, Ma Y, Wang S, Feng Z, Niu X, Cai C, Kong Z, Zhang T, Guo W (2015) GhCFE1A, a dynamic linker between the ER network and actin cytoskeleton, plays an important role in cotton fibre cell initiation and elongation. J Exp Bot 66:1877–1889
Mathur J, Hulskamp M (2002) Microtubules and microfilaments in cell morphogenesis in higher plants. Curr Biol 12:R669–R676
Miller DD, De Ruijter NCA, Bisseling T, Emons AMC (1999) The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharides as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J 17:141–154
Ng DW, Lu J, Chen ZJ (2012) Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol 15:154–161
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin D, Llewellyn D, Showmaker KC, Shu S, Udall J, Yoo MJ, Byers R, Chen W, Doron-Faigenboim A, Duke MV, Gong L, Grimwood J, Grover C, Grupp K, Hu G, Lee TH, Li J, Lin L, Liu T, Marler BS, Page JT, Roberts AW, Romanel E, Sanders WS, Szadkowski E, Tan X, Tang H, Xu C, Wang J, Wang Z, Zhang D, Zhang L, Ashrafi H, Bedon F, Bowers JE, Brubaker CL, Chee PW, Das S, Gingle AR, Haigler CH, Harker D, Hoffmann LV, Hovav R, Jones DC, Lemke C, Mansoor S, ur Rahman M, Rainville LN, Rambani A, Reddy UK, Rong JK, Saranga Y, Scheffler BE, Scheffler JA, Stelly DM, Triplett BA, Van Deynze A, Vaslin MF, Waghmare VN, Walford SA, Wright RJ, Zaki EA, Zhang T, Dennis ES, Mayer KF, Peterson DG, Rokhsar DS, Wang X, Schmutz J (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Ruan YL (2007) Goldacre paper: rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct Plant Biol 34:1–10
Ruan YL, Llewellyn DJ, Furbank RT (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13:47–60
Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116:1539–1549
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Van Ooijen JW, Voorrips RE (2001) JoinMap® Version 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 3:t4995
Wang HY, Wang J, Gao P, Jiao GL, Zhao PM, Li Y, Wang GL, Xia GX (2009) Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol J 7:13–23
Wang H, Guo Y, Lv F, Zhu H, Wu S, Jiang Y, Li F, Zhou B, Guo W, Zhang T (2010a) The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol Biol 72:397–406
Wang J, Wang HY, Zhao PM, Han LB, Jiao GL, Zheng YY, Huang SJ, Xia GX (2010b) Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol 51:1276–1290
Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T (2012) Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24:4012–4025
Wendel JF (1989) New World tetraploid cottons contain Old World cytoplasm. In Proc Natl Acad Sci USA 86:4132–4136
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Yamamoto E, Baird WV (1998) Three cotton fiber-expressed cDNAs (Accession Nos. AF072404, AF072405, and AF072406) (PGR 98–144). Plant Physiol 117:1525–1527
Yoo MJ, Wendel JF (2014) Comparative evolutionary and developmental dynamics of the cotton (Gossypium hirsutum) fiber transcriptome. PLoS Genet 10:e1004073
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537
Zhou R, Moshgabadi N, Adams KL (2011) Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci USA 108:16122–16127
Acknowledgments
This program was financially supported in part by National Natural Science Foundation of China (31471539), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (010-809001), and Jiangsu Collaborative Innovation Center for Modern Crop Production (No. 10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Additional information
Communicated by D.-B. Zhang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lv, F., Li, P., Zhang, R. et al. Functional divergence of GhCFE5 homoeologs revealed in cotton fiber and Arabidopsis root cell development. Plant Cell Rep 35, 867–881 (2016). https://doi.org/10.1007/s00299-015-1928-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1928-0