Abstract
Key message
DhEFL2, 3 and 4 regulate the flowering of Doritaenopsis . These genes could rescue elf4 - 1 phenotype in Arabidopsis while its overexpression delayed flowering.
Abstract
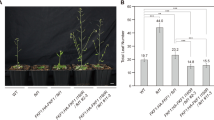
Phalaenopsis are popular floral plants, and studies on orchid flowering genes could help develop off-season cultivars. EARLY FLOWERING 4 (ELF4) of A. thaliana has been shown to be involved in photoperiod perception and circadian regulation. We isolated two members of the ELF4 family from Doritaenopsis hybrid (Doritaenopsis ‘Tinny Tender’ (Doritaenopsis Happy Smile × Happy Valentine)), namely, DhEFL2 and DhEFL3 (DhEFL4 has been previously cloned). Multiple alignment analysis of the deduced amino acid sequences of the three DhEFL homologs showed that DhEFL4 and DhEFL2 are similar with 72 % identical amino acids, whereas DhEFL3 is divergent with 72 % similarity with DhEFL2 and 68 % similarity with DhEFL4. DhEFL3 forms a separate phylogenetic subgroup and is far away from DhEFL2 and DhEFL4. The diurnal expression patterns of DhEFL2, 3, and 4 are similar in the long-day photoperiod conditions; however, in the short-day conditions, DhEFL3 is different from DhEFL2 and 4. For the DhEFL2, 3, and 4 genes, the strongest audience expression organs are the stem, petal and bud, respectively. The ectopic expression of DhEFL2, 3, or 4 in transgenic A. thaliana plants (Ws-2 ecotype) showed novel phenotypes by late flowering and more rosette leaves. The ectopic expression of DhEFL2, 3, or 4 could complement the elf4-1 flowering time and hypocotyl length defects in transgenic A. thaliana elf4-1 mutant plants. These results strongly suggest that DhEFL2, 3, and 4 may be involved in regulation of flower formation and floral induction in Doritaenopsis.






Similar content being viewed by others
Abbreviations
- ELF4 :
-
EARLY FLOWERING 4
- EFL2 :
-
EARLY FLOWERING 4-like2
- EFL3 :
-
EARLY FLOWERING 4-like3
- EFL4 :
-
EARLY FLOWERING 4-like4
- SD:
-
Short day
- LD:
-
Long day
- GI :
-
GIGANTEA
- CCA1:
-
CIRCADIAN CLOCK ASSOCIATED 1
- LHY:
-
LATE ELONGATED HYPOCOTYL
- TOC1 :
-
TIMING OF CAB EXPRESSION 1
- ELF3:
-
EARLY FLOWERING 3
- LUX:
-
LUX ARRHYTHMO
- CaMV:
-
Cauliflower mosaic virus
- EST:
-
Expressed sequence tag
- CO :
-
CONSTANS
- RACE:
-
Rapid amplification of cDNA ends
- ZT:
-
Zeitgeber time
- qRT-PCR:
-
Quantitative real-time PCR
- PCR:
-
Polymerase chain reaction
- MS:
-
Murashige and Skoog
- ORF:
-
Open reading frame
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- WT:
-
Wild type
References
Adeyemo OS, Kolmos E, Tohme J, Chavariaga P, Fregene M, Davis SJ (2011) Identification and characterization of the cassava core-clock gene EARLY FLOWERING 4. Trop Plant Biol 4:117–125
Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293:880–883
Atwood JT (1986) The size of the Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 9:171–186
Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J (2005) Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol 137:969–982
Chao Y-T, Su C-L, Jean W-H, Chen W-C, Chang Y-CA, Shih M-C (2014) Identification and characterization of the microRNA transcriptome of a moth orchid Phalaenopsis aphrodite. Plant Mol Biol 84:529–548
Chen W, Qin Q, Zheng Y, Wang C, Wang S, Zhang C, Zhou M, Cui Y (2015) Overexpression of Doritaenopsis hybrid EARLY FLOWERING 4-like4 gene, DhEFL4, postpones flowering in transgenic Arabidopsis. Plant Mol Biol Report in press
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui Y-Y, Pandey DM, Hahn E-J, Paek K-Y (2004) Effect of drought on physiological aspects of Crassulacean acid metabolism in Doritaenopsis. Plant Sci 167:1219–1226
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 1:19–21
Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419:74–77
Hsiao Y-Y, Chen Y-W, Huang S-C, Pan Z-J, Fu C-H, Chen W-H, Tsai W-C, Chen H-H (2011) Gene discovery using next-generation pyrosequencing to develop ESTs for Phalaenopsis orchids. BMC Genom 12:360
Hsu C-C, Chung Y-L, Chen T-C, Lee Y-L, Kuo Y-T, Tsai W-C, Hsiao Y-Y, Chen Y-W, Wu W-L, Chen H-H (2011) An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol 11:3
Khanna R, Kikis EA, Quail PH (2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133:1530–1538
Kiselev KV, Turlenko AV, Zhuravlev YN (2010) Structure and expression profiling of a novel calcium-dependent protein kinase gene PgCDPK1a in roots, leaves, and cell cultures of Panax ginseng. Plant Cell Tissue Organ Cult (PCTOC) 103:197–204
Kolmos E, Davis SJ (2007) ELF4 as a central gene in the circadian clock. Plant Signal Behav 2:370–372
Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ (2009) Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J 3:350–366
McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144:391–401
Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48:110–121
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402
Oliverio KA, Crepy M, Martin-Tryon EL, Milich R, Harmer SL, Putterill J, Yanovsky MJ, Casal JJ (2007) GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol 144:495–502
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857
Qin Q, Kaas Q, Zhang C, Zhou L, Luo X, Zhou M, Sun X, Zhang L, Paek K-Y, Cui Y (2012) The cold awakening of Doritaenopsis ‘Tinny Tender’ orchid flowers: the role of leaves in cold-induced bud dormancy release. J Plant Growth Regul 31:139–155
Sun X, Qin Q, Zhang J, Zhang C, Zhou M, Paek KY, Cui Y (2012) Isolation and characterization of the FVE gene of a Doritaenopsis hybrid involved in the regulation of flowering. Plant Growth Regul 68:77–86
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang Z-Y, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217
Xuan N, Jin Y, Zhang H, Xie Y, Liu Y, Wang G (2011) A putative maize zinc-finger protein gene, ZmAN13, participates in abiotic stress response. Plant Cell Tissue Organ Cult (PCTOC) 107:101–112
Zakizadeh H, Stummann BM, Lütken H, Müller R (2010) Isolation and characterization of four somatic embryogenesis receptor-like kinase (RhSERK) genes from miniature potted rose (Rosa hybrida cv. Linda). Plant Cell Tissue Organ Cult (PCTOC) 101:331–338
Zhang J-X, Wu K-L, Tian L-N, Zeng S-J, Duan J (2011) Cloning and characterization of a novel CONSTANS-like gene from Phalaenopsis hybrida. Acta Physiol Plant 33:409–417
Acknowledgments
We thank Dr. Zhi John Lu, MOE Key of Bioinformatics, School of Life Science, Tsinghua University, for the useful suggestions on improving the manuscript, and Mr. B. B. Shi, School of Medicine, Tsinghua University, for the useful suggestions on drafting figures.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflicts of interest declared.
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 30771762, 31170658, and 31301811) and the Science & Technology Key Project of Zhejiang Province (Grant No. 2012C12909-11).
Additional information
Communicated by P. Lakshmanan.
The nucleotide sequence reported in this paper has been submitted to GenBank with accession numbers KP728997 (DhEFL2 mRNA), KP728998 (DhEFL3 mRNA).
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2015_1848_MOESM1_ESM.tif
Supplementary material 1 (TIFF 14597 kb) Online Resource 1 Nucleotide and deduced amino acid sequences of DhEFL2 and 3 cDNAs. The DUF1313 superfamily region is indicated with the parenthesis in bold
299_2015_1848_MOESM2_ESM.docx
Supplementary material 2 (DOCX 72 kb) Online Resource 2 Sequence information on alignment of the DhEFL2, 3, 4 amino acid sequences with ELF4 and ELF4-like from different plant species.
299_2015_1848_MOESM3_ESM.docx
Supplementary material 3 (DOCX 16 kb) Online Resource 3 Sequence information on phylogenetic analysis of the DhEFL2, 3, and 4 encoded amino acid sequences from different plant species
Rights and permissions
About this article
Cite this article
Chen, W., Qin, Q., Zhang, C. et al. DhEFL2, 3 and 4, the three EARLY FLOWERING4-like genes in a Doritaenopsis hybrid regulate floral transition. Plant Cell Rep 34, 2027–2041 (2015). https://doi.org/10.1007/s00299-015-1848-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1848-z