Abstract
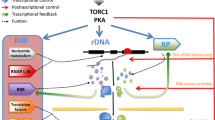
In Saccharomyces cerevisiae, the large majority of the genes coding for cytoplasmic ribosomal proteins (RPs) depend on the general regulatory factor Rap1 for their transcription, but a small cohort of them relies on Abf1 regulatory activity. A recent study showed that unlike Rap1, whose association with RP gene promoters is not affected by environmental changes causing RP gene repression/reactivation, Abf1 association with both RP gene and ribosome biogenesis (Ribi) gene promoters dynamically responds to changes in growth conditions. This observation changes the paradigm of general regulatory factors as relatively static DNA-binding proteins constitutively bound to highly active promoters, and point to Abf1, which binds hundreds of non-RPG promoters within the yeast genome, as a possible key regulatory switch in nutrient- and stress-dependent transcriptional modulation. Moreover, the frequent presence of Abf1 binding sites in the promoters of mitochondrial RP genes evokes the possibility that Abf1 might orchestrate still unexplored levels of co-regulation involving growth-related gene networks in yeast cells.
Similar content being viewed by others
References
Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteom: MCP 7:1389–1396
Azad GK, Tomar RS (2016) The multifunctional transcription factor Rap1: a regulator of yeast physiology. Front Biosci 21:918–930
Bhattacharya A, Warner JR (2008) Tbf1 or not Tbf1? Mol Cell 29:537–538
Bosio MC, Fermi B, Spagnoli G, Levati E, Rubbi L, Ferrari R, Pellegrini M, Dieci G et al (2016) Abf1 and other general regulatory factors control ribosome biogenesis gene expression in budding yeast (Submitted)
Brigati C, Kurtz S, Balderes D, Vidali G, Shore D (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol 13:1306–1314
Chang DT, Huang CY, Wu CY, Wu WS (2011) YPA: an integrated repository of promoter features in Saccharomyces cerevisiae. Nucl Acids Res 39:D647–D652
Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD (1990) A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev 4:503–514
Fermi B, Bosio MC, Dieci G (2016) Promoter architecture and transcriptional regulation of Abf1-dependent ribosomal protein genes in Saccharomyces cerevisiae. Nucl Acids Res. doi:10.1093/nar/gkw194
Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18:2522–2537
Francesconi SC, Eisenberg S (1991) The multifunctional protein OBF1 is phosphorylated at serine and threonine residues in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 88:4089–4093
Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH (2011) Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucl Acids Res 39:2032–2044
Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9:383–387
Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA (2004) Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104
Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458
Ho YH, Gasch AP (2015) Exploiting the yeast stress activated signaling network to inform on stress biology and disease signaling. Curr Genet 61:503–511
Ju QD, Morrow BE, Warner JR (1990) REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol 10:5226–5234
Lascaris RF, Mager WH, Planta RJ (1999) DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics 15:267–277
Mager WH, Planta RJ (1990) Multifunctional DNA-binding proteins mediate concerted transcription activation of yeast ribosomal protein genes. Biochim Biophys Acta 1050:351–355
Morrow BE, Ju Q, Warner JR (1990) Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem 265:20778–20783
Oliveira AP, Ludwig C, Zampieri M, Weisser H, Aebersold R, Sauer U (2015) Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci Signal 8:rs4
Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, Schultz P (2010) TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465:956–960
Paul E, Tirosh I, Lai W, Buck MJ, Palumbo MJ, Morse RH (2015) Chromatin mediation of a transcriptional memory effect in yeast. G3 5:829–838
Raychaudhuri S, Byers R, Upton T, Eisenberg S (1997) Functional analysis of a replication origin from Saccharomyces cerevisiae: identification of a new replication enhancer. Nucl Acids Res 25:5057–5064
Reed SH, Akiyama M, Stillman B, Friedberg EC (1999) Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev 13:3052–3058
Schlecht U, Erb I, Demougin P, Robine N, Borde V, van Nimwegen E, Nicolas A, Primig M (2008) Genome-wide expression profiling, in vivo DNA binding analysis, and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration, and sporulation in yeast. Mol Biol Cell 19:2193–2207
Shore D (1994) RAP1: a protean regulator in yeast. Trends Genet 10:408–412
Silve S, Rhode PR, Coll B, Campbell J, Poyton RO (1992) ABF1 is a phosphoprotein and plays a role in carbon source control of COX6 transcription in Saccharomyces cerevisiae. Mol Cell Biol 12:4197–4208
Soontorngun N (2016) Reprogramming of nonfermentative metabolism by stress responsive transcription factors in the yeast Saccharomyces cerevisiae. Curr Genet. doi:10.1007/s00294-016-0609-z
Woo DK, Phang TL, Trawick JD, Poyton RO (2009) Multiple pathways of mitochondrial-nuclear communication in yeast: intergenomic signaling involves ABF1 and affects a different set of genes than retrograde regulation. Biochim Biophys Acta 1789:135–145
Yarragudi A, Parfrey LW, Morse RH (2007) Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucl Acids Res 35:193–202
Yu S, Smirnova JB, Friedberg EC, Stillman B, Akiyama M, Owen-Hughes T, Waters R, Reed SH (2009) ABF1-binding sites promote efficient global genome nucleotide excision repair. J Biol Chem 284:966–973
Acknowledgments
We thank Paola Frigeri (University of Parma) for help with MRP promoter analysis. This work was supported by a Grant from the Italian Ministry of Education, University and Research (MIUR, PRIN 2009 to G.D.) and Italian Association for Cancer Research (AIRC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Fermi, B., Bosio, M.C. & Dieci, G. Multiple roles of the general regulatory factor Abf1 in yeast ribosome biogenesis. Curr Genet 63, 65–68 (2017). https://doi.org/10.1007/s00294-016-0621-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0621-3