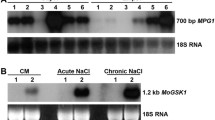
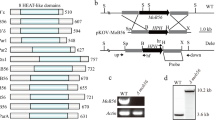
Abstract
Protein phosphatase 2A is a subgroup of widely conserved serine/threonine phosphatases and plays diverse roles in transcription, translation, differentiation, cell cycle, and signal transduction in many organisms. However, its roles in biotrophic and hemi-biotrophic phytopathogenic fungi remain to be investigated. In this study, we isolated an insertional mutant of the rice blast fungus Magnaporthe oryzae that was defective in vegetative hyphal growth. In the mutant, the T-DNA fragment was found to be inserted in the promoter region of a putative serine/threonine protein phosphatase 2A catalytic subunit (PP2Ac) gene MoPPG1. Deletion of MoPPG1 leads to severe defects in vegetative hyphal growth and conidiation. Conidia of the ∆Moppg1 null mutants were misshaped, and most of them were two-celled. The deletion mutants of MoPPG1 did not penetrate into host plant cells and failed to cause any disease lesions on rice leaves. Interestingly, significant reduction was found in the ∆Moppg1 null mutants in expression levels of several Rho GTPase family genes including MgCDC42, MgRHO3, and MgRAC1, which were important for pathogenesis of M. oryzae. Taken together, our results indicated that PP2Ac plays vital roles in asexual development and plant infection by regulating Rho GTPases in the rice blast fungus and perhaps other plant pathogenic fungi.






Similar content being viewed by others
References
Blacketer MJ, Koehler CM, Coats SG, Myers AM, Madaule P (1993) Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein. Mol Cell Biol 13:5567–5581
Chen J, Zheng W, Zheng S, Zhang D, Sang W, Chen X, Li G, Lu G, Wang Z (2008) Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog 4:e1000202
Chen XL, Yang J, Peng YL (2011) Large scale insertional mutagenesis by Agrobacterium tumefaciens-mediated transformation. Methods Mol Biol 722:213–224
Choi YE, Shim WB (2008) Functional characterization of Fusarium verticillioides CPP1, a gene encoding a putative protein phosphatase 2A catalytic subunit. Microbiology 154:326–336
Choi JH, Kim YS, Lee YH (2009) Functional analysis of MCNA, a gene encoding a catalytic subunit of calcineurin, in rice blast fungus Magnaporthe oryzae. J Microbiol Biotechnol 19:11–16
Cohen PTW (2003) Overview of protein serine/threonine phosphatases. Top Curr Genet 5:1–20
Dagdas YF, Yoshino K, Dagdas G, Ryder LS, Bielska E, Steinberg G, Talbot NJ (2012) Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336:1590–1595
de Jong JC, McCormack BJ, Smirnoff N, Talbot NJ (1997) Glycerol generates turgor in rice blast. Nature 389:244–245
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan HQ, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li WX, Hardling M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe oryzae. Nature 434:980–986
Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430
Ebbole DJ (2007) Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 45:437–456
Erental A, Harel A, Yarden O (2007) Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum. Mol Plant Microbe Interact 20:944–954
Gallego M, Virshup DM (2005) Protein serine/threonine phosphatases: life, death, and sleeping. Curr Opin Cell Biol 17:197–202
Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305–308
Jaffe AB, Hall A (2005) Rho GTPases: biochemistry and biology. Ann Rev Cell Dev Biol 21:247–269
Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem J 353:417–439
Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S, Choi J, Park JY, Yi M, Yang S, Kwon MJ, Han SS, Kim BR, Khang CH, Park B, Lim SE, Jung K, Kong S, Karunakaran M, Oh HS, Kim H, Kim S, Park J, Kang S, Choi WB, Kang S, Lee YH (2007) Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39:561–565
Jiang Y (2006) Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:440–449
Johnson DI (1999) Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev 63:54–105
Kang Z, Zingen-Sell I, Buchenauer H (2005) Infection of wheat spikes by Fusarium avenaceum and alterations of cell wall components in the infected tissue. Eur J Plant Pathol 111:19–28
Kankanala P, Czymmek K, Valent B (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19:706–724
Kinoshita N, Ohkura H, Yanagida M (1990) Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fusion yeast cell division cycle. Cell 63:405–415
Kong LA, Yang J, Li GT, Qi LL, Zhang Y, Wang CF, Zhao WS, Xu JR, Peng YL (2012) Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathogens 8:1002526
Kosmidou E, Lunness P, Doonan JH (2001) A type 2A protein phosphatase gene from Aspergillus nidulans is involved in hyphal morphogenesis. Curr Genet 39:25–34
Lechward K, Awotunde OS, Swiatek W, Muszynska G (2001) Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim Pol 48:921–933
Li G, Zhou X, Xu JR (2012) Genetic control of infection-related development in Magnaporthe oryzae. Curr Opin Microbiol. doi:10.1016/j.mib.2012.09.004
Lin FC, Arndt KT (1995) The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and entry into meiosis. EMBO J 14:2745–2759
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–462
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Mayer-Yakel RE, Hemmings BA (1994) Protein phosphatase 2A—a ‘menage a trois’. Trends Cell Biol 4:287–291
Mehrabi R, Ding SL, Xu JR (2008) The MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell 7:791–799
Orgad S, Brewis ND, Alphey L, Axton JM, Dudai Y, Cohen PTW (1990) The structure of protein phosphatase 2A is as highly conserved as that of protein phosphatase 1. FEBS Lett 275:44–48
Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR (2004) Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol 53:1695–1707
Peng YL, Shishiyama J (1988) Temporal sequence of cytological events in rice leaves infected with Pyricularia oryzae. Can J Bot 66:730–735
Ronne H, Carlberg M, Hu GZ, Nehlin JO (1991) Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol Cell Biol 11:4876–4884
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Saunders DGO, Aves SJ, Talbot NJ (2010) Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22:497–507
Skamnioti P, Gurr SJ (2007) Magnaporthe grisea cutinase 2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 19:2674–2689
Talbot NJ, Kershaw MJ, Wakley GE, de Vries OMH, Wessels JGH, Hamer JE (1996) MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8:985–999
Tanabe O, Hirata D, Usui H, Nishito Y, Miyakawa T, Igarashi K, Takeda M (2001) Fission yeast homologues of the B′ subunit of protein phosphatase 2A: multiple roles in mitotic cell division and functional interaction with calcineurin. Genes Cells 6:455–473
Thines E, Weber RWS, Talbot NJ (2000) MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703–1718
Tsuji G, Fujii S, Fujihara N, Hirose C, Tsuge S, Shiraishi T, Kubo Y (2003) Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J Gen Plant Pathol 69:230–239
Valent B (1990) Rice blast as a model system for plant pathology. Phytopathol 80:33–36
Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ (2006) Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312:580–583
Wang H, Jiang Y (2003) The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol Cell Biol 23:3116–3125
Wang H, Wang X, Jiang Y (2003) Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol Biol Cell 14:4342–4351
Wilson RA, Talbot NJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nature Rev Microbiol 7:185–195
Xu JR, Hamer JE (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe oryzae. Genes Dev 10:2696–2706
Xu JR, Peng YL, Dickman MB, Sharon A (2006) The dawn of fungal pathogen genomics. Annu Rev Phytopathol 44:337–366
Xu JR, Zhao XH, Dean RA (2007) From genes to genomics: a new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv Genet 57:175–218
Xue M, Yang J, Li Z, Hu S, Yao N, Dean RA, Zhao W, Shen M, Zhang H, Li C, Liu L, Cao L, Xu X, Xing Y, Hsiang T, Zhang Z, Xu JR, Peng YL (2012) Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet 8:e1002869
Yang J, Zhao XY, Sun J, Kang ZS, Ding SL, Xu JR, Peng YL (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant Microbe Interact 23:112–123
Yang J, Kong LA, Chen XL, Wang DW, Qi LL, Zhao WS, Zhang Y, Liu XZ, Peng YL (2012) A carnitine-acylcarnitine carrier protein, MoCrc1, is essential for pathogenicity in Magnaporthe oryzae. Curr Genet 58:139–148
Yatzkan E, Szoor B, Feher Z, Dombradi V, Yarden O (1998) Protein phosphatase 2A is involved in hyphal growth of Neurospora crassa. Mol Gen Genet 259:523–531
Zhao X, Mehrabi R, Xu JR (2007) MAP kinase pathways and fungal pathogenesis. Eukaryot Cell 10:1701–1714
Zheng W, Chen J, Zheng S, Liu L, Liu S, Lu G, Zhou J, Wang Z (2006) Rho proteins and the expression module of their encoding genes in Magnaporthe grisea. Acta Phytopathol Sinica 36:328–336
Zheng W, Chen J, Liu W, Zheng S, Zhou J, Lu G, Wang Z (2007) A Rho3 homolog is essential for appressorium development and pathogenicity of Magnaporthe grisea. Eukaryot Cell 6:2240–2250
Zheng W, Zhao Z, Chen J, Liu W, Ke H, Zhou J, Lu G, Darvill AG, Albersheim P, Wu S, Wang Z (2009) A Cdc42 ortholog is required for penetration and virulence of Magnaporthe grisea. Fungal Genet Biol 46:450–460
Acknowledgments
We thank Dr. Wensheng Zhao and Yan Zhang at China Agricultural University for helpful discussions. This work was supported by a grant from the National Fundamental Basic Research program (2012CB114002) to Y. -L. Peng of the Ministry of Sciences and Technology, China.
Conflict of interest
We declared that no conflict of interest exists.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Borkovich.
Y. Du and Y. Shi contributed equally to this study. Nucleotide sequence data reported is available in the GenBank database under the accession number AEK98536 (MoPPG1).
Electronic supplementary material
Below is the link to the electronic supplementary material.
294_2012_385_MOESM1_ESM.tif
Fig. S1 Alignment and phylogenetic tree of MoPpg1 with its orthologs. The alignment was generated with the ClustalX and ESPript programs using protein sequences MoPpg1 (Magnaporthe oryzae), EAA33034 (Neurospora crassa), EAK83067 (Ustilago maydis), Cpp1 (Fusarium verticillioides), CAK43262 (Aspergillus nidulans), SPAC22H10.04 (Schizosaccharomyces pombe), Ppg1 (Saccharomyces cerevisiae), pph1 (Sclerotinia sclerotiorium), PP2A-4 (Arabidopsis thaliana), PP2A-1 (Oryza sativa), PPH-4.1 (Caenorhabditis elegans), PP2AA (Homo sapiens). Identical residues among the sequences are marked in white within red shadow, and similar amino acids are marked in red within blue rectangle box. (TIFF 1324 kb)
Rights and permissions
About this article
Cite this article
Du, Y., Shi, Y., Yang, J. et al. A serine/threonine-protein phosphatase PP2A catalytic subunit is essential for asexual development and plant infection in Magnaporthe oryzae . Curr Genet 59, 33–41 (2013). https://doi.org/10.1007/s00294-012-0385-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-012-0385-3