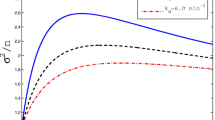
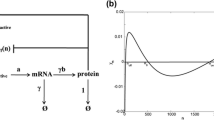
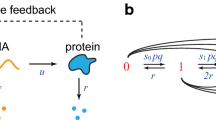
Abstract
We consider the dynamics of a population of organisms containing two mutually inhibitory gene regulatory networks, that can result in a bistable switch-like behaviour. We completely characterize their local and global dynamics in the absence of any noise, and then go on to consider the effects of either noise coming from bursting (transcription or translation), or Gaussian noise in molecular degradation rates when there is a dominant slow variable in the system. We show analytically how the steady state distribution in the population can range from a single unimodal distribution through a bimodal distribution and give the explicit analytic form for the invariant stationary density which is globally asymptotically stable. Rather remarkably, the behaviour of the stationary density with respect to the parameters characterizing the molecular behaviour of the bistable switch is qualitatively identical in the presence of noise coming from bursting as well as in the presence of Gaussian noise in the degradation rate. This implies that one cannot distinguish between either the dominant source or nature of noise based on the stationary molecular distribution in a population of cells. We finally show that the switch model with bursting but two dominant slow genes has an asymptotically stable stationary density.







Similar content being viewed by others
References
Angeli D, Ferrell JE, Sontag ED (2004) Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc Natl Acad Sci USA 101(7):1822–1827
Artyomov MN, Das J, Kardar M, Chakraborty AK (2007) Purely stochastic binary decisions in cell signaling models without underlying deterministic bistabilities. Proc Natl Acad Sci USA 104(48):18958–18963
Bishop LM, Qian H (2010) Stochastic bistability and bifurcation in a mesoscopic signaling system with autocatalytic kinase. Biophys J 98(1):1–11
Bokes P, King JR, Wood AT, Loose M (2013) Transcriptional bursting diversifies the behaviour of a toggle switch: hybrid simulation of stochastic gene expression. Bull Math Biol 75(2):351–371
Cai L, Friedman N, Xie X (2006) Stochastic protein expression in individual cells at the single molecule level. Nature 440:358–362
Caravagna G, Mauri G, d’Onofrio A (2013) The interplay of intrinsic and extrinsic bounded noises in biomolecular networks. PLoS One 8(2):e51,174
Cherry J, Adler F (2000) How to make a biological switch. J Theoret Biol 203:117–133
Chubb J, Trcek T, Shenoy S, Singer R (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16:1018–1025
Davis M (1993) Monographs on statistics and applied probability, vol 49, Markov models and optimization. Chapman & Hall, London
Eldar A, Elowitz MB (2010) Functional roles for noise in genetic circuits. Nature 467(7312):167–173
Ferrell JE (2002) Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 14(2):140–148
Gardner T, Cantor C, Collins J (2000) Construction of a genetic toggle switch in \(Escherichia\; coli\). Nature 403:339–342
Gillespie D (2000) The chemical Langvin equation. J Chem Phys 113:297–306
Golding I, Paulsson J, Zawilski S, Cox E (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123:1025–1036
Goodwin BC (1965) Oscillatory behavior in enzymatic control processes. Adv Enzym Regulat 3:425–428 (IN1–IN2, 429–430, IN3–IN6, 431–437). doi:10.1016/0065-2571(65)90067-1
Griffith J (1968a) Mathematics of cellular control processes. I. Negative feedback to one gene. J Theor Biol 20:202–208
Griffith J (1968b) Mathematics of cellular control processes. II. Positive feedback to one gene. J Theor Biol 20:209–216
Grigorov L, Polyakova M, Chernavskil D (1967) Model investigation of trigger schemes and the differentiation process (in Russian). Mol Biol 1(3):410–418
Haken H (1983) Springer series in synergetics, vol 1, 3rd edn., Synergetics: an introduction. Springer, Berlin
Hasty J, Isaacs F, Dolnik M, McMillen D, Collins JJ (2001) Designer gene networks: towards fundamental cellular control. Chaos 11(1):207–220
Huang D, Holtz WJ, Maharbiz MM (2012) A genetic bistable switch utilizing nonlinear protein degradation. J Biol Eng 6(1):9
Huang L, Yuan Z, Liu P, Zhou T (2015) Effects of promoter leakage on dynamics of gene expression. BMC Syst Biol 9:16
Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356
Kepler T, Elston T (2001) Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophy J 81:3116–3136
Lasota A, Mackey MC (1994) Chaos, fractals, and noise. Applied Mathematical Sciences, vol 97. Springer, New York
Mackey MC, Tyran-Kamińska M (2008) Dynamics and density evolution in piecewise deterministic growth processes. Ann Polon Math 94:111–129
Mackey MC, Tyran-Kamińska M, Yvinec R (2011) Molecular distributions in gene regulatory dynamics. J Theor Biol 274:84–96
Monod J, Jacob F (1961) Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol 26:389–401
Morelli MJ, Allen RJ, Tanase-Nicola S, ten Wolde PR (2008a) Eliminating fast reactions in stochastic simulations of biochemical networks: a bistable genetic switch. J Chem Phys 128(4):045105
Morelli MJ, Tanase-Nicola S, Allen RJ, ten Wolde PR (2008b) Reaction coordinates for the flipping of genetic switches. Biophys J 94(9):3413–3423
Ochab-Marcinek A, Tabaka M (2015) Transcriptional leakage versus noise: a simple mechanism of conversion between binary and graded response in autoregulated genes. Phys Rev E Stat Nonlinear Soft Matter Phys 91(1):012704
Othmer H (1976) The qualitative dynamics of a class of biochemical control circuits. J Math Biol 3:53–78
Pichór K, Rudnicki R (2000) Continuous Markov semigroups and stability of transport equations. J Math Anal Appl 249:668–685
Polynikis A, Hogan S, di Bernardo M (2009) Comparing differeent ODE modelling approaches for gene regulatory networks. J Theor Biol 261:511–530
Ptashne M (1986) A genetic switch: gene control and phage lambda. Cell Press, Cambridge
Qian H, Shi PZ, Xing J (2009) Stochastic bifurcation, slow fluctuations, and bistability as an origin of biochemical complexity. Phys Chem Chem Phys 11(24):4861–4870
Raj A, Peskin C, Tranchina D, Vargas D, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4:1707–1719
Rudnicki R, Pichór K, Tyran-Kamińska M (2002) Markov semigroups and their applications. In: Dynamics of dissipation. Lectures notes in physics, vol 597. Springer, Berlin, pp 215–238
Samoilov M, Plyasunov S, Arkin AP (2005) Stochastic amplification and signaling in enzymatic futile cycles through noise-induced bistability with oscillations. Proc Natl Acad Sci USA 102(7):2310–2315
Selgrade J (1979) Mathematical analysis of a cellular control process with positive feedback. SIAM J Appl Math 36:219–229
Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U (2006) Variability and memory of protein levels in human cells. Nature 444:643–646
Smith H (1995) Mathematical surveys and monographs, vol 41, Monotone dynamical systems. American Mathematical Society, Providence
Strasser M, Theis FJ, Marr C (2012) Stability and multiattractor dynamics of a toggle switch based on a two-stage model of stochastic gene expression. Biophys J 102(1):19–29
Tyran-Kamińska M (2009) Substochastic semigroups and densities of piecewise deterministic Markov processes. J Math Anal Appl 357:385–402
Tyson JJ, Chen KC, Novak B (2003) Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol 15(2):221–231
Vellela M, Qian H (2009) Stochastic dynamics and non-equilibrium thermodynamics of a bistable chemical system: the Schlgl model revisited. J R Soc Interface 6(39):925–940
Waldherr S, Wu J, Allgower F (2010) Bridging time scales in cellular decision making with a stochastic bistable switch. BMC Syst Biol 4:108
Wang J, Zhang J, Yuan Z, Zhou T (2007) Noise-induced switches in network systems of the genetic toggle switch. BMC Syst Biol 1:50
Yu J, Xiao J, Ren X, Lao K, Xie X (2006) Probing gene expression in live cells, one protein molecule at a time. Science 311:1600–1603
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council (NSERC, Canada) and the Polish NCN grant no 2014/13/B/ST1/00224. We are grateful to Marc Roussel (Lethbridge), Romain Yvinec (Tours) and Changjing Zhuge (Tsinghua University, Beijing) for helpful comments on this problem. We are especially indebted to the referees and the Associate Editor for their comments that have materially improved this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mackey, M.C., Tyran-Kamińska, M. The limiting dynamics of a bistable molecular switch with and without noise. J. Math. Biol. 73, 367–395 (2016). https://doi.org/10.1007/s00285-015-0949-1
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-015-0949-1