Abstract
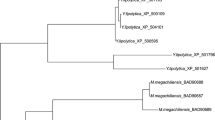
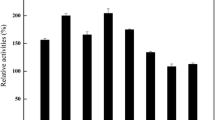
The purpose of the present investigation was to produce erythritol by Yarrowia lipolytica mutant without any by-products. Mutants of Y. lipolytica were generated by ultra-violet for enhancing erythrose reductase (ER) activity and erythritol production. The mutants showing the highest ER activity were screened by triphenyl tetrazolium chloride agar plate assay. Productivity of samples was analyzed by thin-layer chromatography and high-performance liquid chromatography equipped with the refractive index detector. One of the mutants named as mutant 49 gave maximum erythritol production without any other by-products (particularly glycerol). Erythritol production and specific ER activity in mutant 49 increased to 1.65 and 1.47 times, respectively, in comparison with wild-type strain. The ER gene of wild and mutant strains was sequenced and analyzed. A general comparison of wild and mutant gene sequences showed the replacement of Asp270 with Glu270 in ER protein. In order to enhance erythritol production, we used a three component-three level-one response Box–Behnken of response surface methodology model. The optimum medium composition for erythritol production was found to be (g/l) glucose 279.49, ammonium sulfate 9.28, and pH 5.41 with 39.76 erythritol production.









Similar content being viewed by others
References
Abe S, Morioka S (1999) Method of producing erythritol. United State patent 5902739
Biro JC (2008) Correlation between nucleotide composition and folding energy of coding sequences with special attention to wobble bases. Theor Biol Med Model 5:14–22
Bradford MM (1976) A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Care S, Bignon C, Pelissier MC, Blanc E, Canard B, Coutard B (2008) The translation of recombinant proteins in E. coli can be improved by in silico generating and screening random libraries of a 270/+96 mRNA region with respect to the translation initiation codon. Nucleic Acids Res 36:1–6
Ghezelbash GR, Nahvi I, Rabani M (2012) Study of polyols production by Yarrowia lipolytica in batch culture and optimization of growth condition for maximum production. Jundishapur J Microbiol 5:546–549
Hajny GJ, Smith JH, Garver JC (1964) Erythritol production by a yeast-like fungus. Appl Microbiol 12:240–246
Heussen C, Dowdle EB (1980) Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulphate and copolymerized substrates. Anal Biochem 102:196–202
Hirata Y, Igarashi K, Ezaki S, Atomi H, Imanaka T (1999) High-level production of erythritol by strain 618A-01 isolated from pollen. J Biosci Bioeng 87:630–635
Hoffman CS (1997) Current protocols in molecular biology. Wiley, New York
Kim SY, Lee KH, Kim JH, Oh DK (1997) Erythritol production by controlling osmotic pressure in Trigonopsis variabilis. Biotechnol Lett 19:727–729
Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13–37
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Laxman SS, Ramchandra VG, Bhalchandra KV, Karthik N (2011) Strain improvement and statistical media optimization for enhanced erythritol production with minimal by-products from Candida magnoliae mutant R23. Biochem Eng J 55:92–100
Lee DH, Lee YJ, Ryu YW, Seo JH (2010) Molecular cloning and biochemical characterization of a novel erythrose reductase from Candida magnoliae JH110. Microb Cell Fact 9:43–55
Lee JK, Ha SJ, Kim SY, Oh DK (2001) Increased erythritol production in Torula sp. with inositol and phytic acid. Biotechnol Lett 23:497–500
Lee JK, Hong KW, Kim SY (2003) Purification and properties of a NADPH-dependent erythrose reductase from the newly isolated Torula coralline. Biotechnol Prog 19:495–500
Lee JK, Kim SY, Ryu YW, Seo JH, Kim JH (2003) Purification and characterization of a novel erythrose reductase from Candida magnoliae. Appl Environ Microbiol 69:3710–3718
Lee KJ, Lim JY (2003) Optimized condition for high erythritol production by Penicillium sp. KJ-UV29, mutant of Penicillium sp. KJg81. Biotechnol Bioprocess Eng 8:173–178
Lin SJ, Wen CY, Liau JC, Chu WS (2001) Screening and production of erythritol by newly isolated osmophilic yeast-like fungi. Process Biochem 36:1249–1258
Lin SJ, Wen CY, Wang PM, Huang JC, Wei CL, Chang JW, Chu WS (2010) High-level production of erythritol by mutants of osmophilic Moniliella sp. Process Biochem 45:973–979
Park EH, Lee HY, Ryu YW, Seo JH, Kim MD (2011) Role of osmotic and salt stress in the expression of erythrose reductase in Candida magnoliae. J Microbiol Biotechnol 10:1064–1068
Park JB, Seo BC, Kim JR, Park YK (1998) Production of erythritol in fed-batch cultures of Trichosporon sp. J Ferment Bioeng 86:577–580
Park YC, Lee DY, Lee DH, Kim HJ, Ryu YW, Seo JH (2005) Proteomics and physiology of erythritol-producing strains. J Chromatogr B 815:251–260
Patil SV, Jayaraman VK, Kulkarni BD (2002) Optimization of media by evolutionary algorithms for production of polyols. Appl Biochem Biotechnol 102:119–128
Rymowicz W, Rywinska A, Marcinkiewicz M (2009) High-yield production of erythritol from raw glycerol in fed-batch cultures of Yarrowia lipolytica. Biotechnol Lett 31:377–380
Shukla P, Garai D, Zafar M, Gupta K, Shrivastava S (2007) Process parameters optimization for lipase production by Rhizopus oryzae kg-10 under submerged fermentation using response surface methodology. J Appl Sci Environ Sanit 2:93–103
Tokuoka K, Ishitani T, Chungz WC (1992) Accumulation of polyols and sugars in some sugar tolerant yeasts. J Gen Appl Microbiol 38:35–46
Tomaszewska L, Rywinska A, Gładkowski W (2012) Production of erythritol and mannitol by Yarrowia lipolytica yeast in media containing glycerol. J Ind Microbiol Biotechnol 39:1333–1343
Acknowledgments
The current study was supported by the grant of Postgraduate Administration Office of the University of Isfahan to Gh. R. Ghezelbash for obtaining Ph.D. degree. In addition, the authors are grateful to Sara Ghashghaei and Vahid Niknezhad from University of Isfahan for the helps with experiments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghezelbash, G.R., Nahvi, I. & Emamzadeh, R. Improvement of Erythrose Reductase Activity, Deletion of By-products and Statistical Media Optimization for Enhanced Erythritol Production from Yarrowia lipolytica Mutant 49. Curr Microbiol 69, 149–157 (2014). https://doi.org/10.1007/s00284-014-0562-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0562-3