Abstract
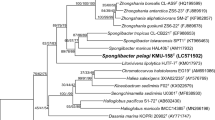
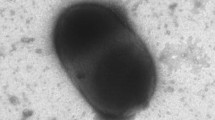
A Gram-negative, rod-shaped, non-spore-forming aerobic bacterium, motile with a single polar flagellum, strain JLT2005T, was isolated from surface seawater collected from the East China Sea and formed ivory white colonies on a rich organic medium. The strain was positive for catalase, oxidase, and urease. It grew in the presence of 0–12 % (w/v) NaCl (optimum 5 %), at 20–35 °C (optimum 25 °C), or at pH 6–10 (optimum pH 9). The major fatty acids (>10 %) were C18:1ω7c, C19:0ω8c cyclo, C16:0, and C18:0. The major polar lipids were phosphatidylglycerol, diphosphatidylglycerol, and five unidentified glycolipids. Ubiquinone-10 and Ubiquinone-11 were present as the major quinones. The DNA G+C content was 74.3 mol%. Phylogenetic analyses based on 16S rRNA gene sequences indicated that strain JLT2005T belongs to the genus Pelagibacterium in the family Hyphomicrobiaceae, class Alphaproteobacteria. The closest neighbors were Pelagibacterium halotolerans B2T (98.7 % similarity) and Pelagibacterium luteolum 1_C16_27T (97.1 % similarity). DNA–DNA relatedness values of strain JLT2005T with P. halotolerans B2T and with P. luteolum 1_C16_27T were 31.6 and 25 %. Evidence from genotypic, chemotaxonomic, and phenotypic data shows that strain JLT2005T represents a novel species of the genus Pelagibacterium, for which the name Pelagibacterium nitratireducens sp. nov is proposed. The type strain is JLT2005T (=CGMCC 1.10829T =JCM 17767T).


Similar content being viewed by others
References
Xu X-W, Huo Y–Y, Wang C-S, Oren A, Cui H-L, Vedler E, Wu M (2011) Pelagibacterium halolerans gen.nov., sp. nov. and Pelagibacterium luteolum sp. nov., novel members of the family Hyphomicrobiaceae. Int J Syst Evol Microbiol 61:1817–1822
Drews G (1983) Mikrobiologisches Praktikum (in German). Springer, Berlin
Yurkov VV, Krieger S, Stackebrandt E, Beatty JT (1999) Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J Bacteriol 181(15):4517–4525
Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC
Dong X-Z, Cai M-Y (2001) Determinative manual for routine bacteriology. Scientific, Beijing
Clarke PH (1953) Hydrogen sulphide production by bacteria. J Gen Microbiol 8:397–407
Mata JA, Martínez-Cánovas J, Quesada E, Béjar V (2002) A detailed phenotypic characterisation of the type strains of Halomonas species. Syst Appl Microbiol 25:360–375
Barritt MM (1936) The intensification of the Voges-Proskauer reaction by the addition of a-naphthol. J Pathol Bacteriol 42:441–445
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Xu X-W, Wu Y-H, Wang C-S, Yang J-Y, Oren A, Wu M (2008) Marinobacter pelagius sp. nov. a moderately halophilic bacterium. Int J Syst Evol Microbiol 58:637–640
Hiraishi A, Ueda Y, Ishihara J (1998) Quinone profiling of bacterial communities in natural and synthetic sewage activated sludge for enhanced phosphate removal. App Environ Microbiol 64:992–998
Romano I, Lama L, Nicolaus B, Poli A, Gambacorta A, Giordano A (2006) Halomonas alkaliphila sp. nov., a novel halotolerant alkaliphilic bacterium isolated from a salt pool in Campania (Italy). J Gen Appl Microbiol 52:339–348
Kates M (1986) Techniques of Lipidology, 2nd edn. Elsevier, Amsterdam
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Johnson JL (1994) Similarity analysis of DNAs. In: P. E. Gerhardt, R. G. Murray, W. A. Wood & N. R. Krieg (eds) Methods for general and molecular bacteriology, pp 655–681
Marmur J, Doty P (1962) Determination of the base composition of DNA from its thermal denaturation temperature. J Mol Biol 5:109–118
De Ley J (1970) Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol 101:738–754
Embley TM (1991) The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol 13:171–174
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Chun J, Lee J-H, Jung Y, Kim M, Kim BK, Lim Y-W (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Acknowledgments
The authors would like to thank Dr. Yanan Wang, Key Laboratory of Microbial Engineering at the Institute of Biology, Henan Academy of Sciences; Dr. Xiaojun Yan, School of Life Science & Biological Engineering, Ningbo University; Dr. Henglin Cui, School of Food & Biological Engineering, Jiangsu University; and Dr. Li Gu, Third Institute of Oceanography, State Oceanic Administration for their valuable assistance. Professor John Hodgkiss is also thanked for his assistance with English. This work was supported by 973 program (2011CB808800), SOA project (201105021), MOST project (2012AA092003), and NSFC (41191021). Dr. Rui Zhang was also supported by NSFC (40906059).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Q., Xu, Y., Liu, K. et al. Pelagibacterium nitratireducens sp.nov., A Marine Alphaproteobacterium Isolated from the East China Sea. Curr Microbiol 66, 450–455 (2013). https://doi.org/10.1007/s00284-012-0299-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0299-9