Abstract
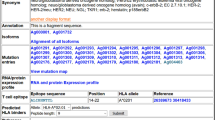
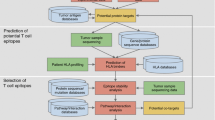
Tumor T cell antigens are both diagnostically and therapeutically valuable molecules. A large number of new peptides are examined as potential tumor epitopes each year, yet there is no infrastructure for storing and accessing the results of these experiments. We have retroactively cataloged more than 1000 tumor peptides from 368 different proteins, and implemented a web-accessible infrastructure for storing and accessing these experimental results. All peptides in TANTIGEN are labeled as one of the four categories: (1) peptides measured in vitro to bind the HLA, but not reported to elicit either in vivo or in vitro T cell response, (2) peptides found to bind the HLA and to elicit an in vitro T cell response, (3) peptides shown to elicit in vivo tumor rejection, and (4) peptides processed and naturally presented as defined by physical detection. In addition to T cell response, we also annotate peptides that are naturally processed HLA binders, e.g., peptides eluted from HLA in mass spectrometry studies. TANTIGEN provides a rich data resource for tumor-associated epitope and neoepitope discovery studies and is freely available at http://cvc.dfci.harvard.edu/tantigen/ or http://projects.met-hilab.org/tadb (mirror).
Similar content being viewed by others
Abbreviations
- BLAST:
-
Basic local alignment search tool
- COSMIC:
-
The catalogue of somatic mutations in cancer
- NCBI:
-
National Center for Biotechnology Information
- ORF:
-
Open reading frame
- TAs:
-
Tumor antigens
- TSAs:
-
Tumor-specific antigens
References
Boon T, Cerottini JC, van den Eynde B et al (1994) Tumor antigens recognized by T lymphocytes. Annu Rev Immunol 12:337–365. doi:10.1146/annurev.iy.12.040194.002005
Olsen L, Campos B, Winther O et al (2014) Tumor antigens as proteogenomic biomarkers in invasive ductal carcinomas. BMC Med Genomics 7(Suppl 3):S2. doi:10.1186/1755-8794-7-S3-S2
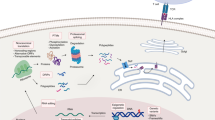
Blum JS, Wearsch PA, Cresswell P (2013) Pathways of antigen processing. Annu Rev Immunol 31:443–473. doi:10.1146/annurev-immunol-032712-095910
Boon T, van der Bruggen P (1996) Human tumor antigens recognized by T lymphocytes. J Exp Med 183:725–729
Restifo NP, Dudley ME, Rosenberg S a (2012) Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol 12:269–281. doi:10.1038/nri3191
Gill S, June CH (2015) Going viral: chimeric antigen receptor T-cell therapy for hematological malignancies. Immunol Rev 263:68–89. doi:10.1111/imr.12243
Srivastava PK (2015) Neoepitopes of Cancers: Looking Back, Looking Ahead. Cancer. Immunol Res 3:969–977. doi:10.1158/2326-6066.CIR-15-0134
Reinhold B, Keskin DB, Reinherz EL (2010) Molecular Detection of Targeted Major Histocompatibility Complex I-Bound Peptides Using a Probabilistic Measure and Nanospray MS(3) on a Hybrid Quadrupole-Linear Ion Trap. Anal Chem 82:9090–9099. doi:10.1021/ac102387t
Reinherz EL, Keskin DB, Reinhold B (2014) Forward vaccinology: CTL targeting based upon physical detection of HLA-bound peptides. Front Immunol 5:418. doi:10.3389/fimmu.2014.00418
Castle JC, Kreiter S, Diekmann J et al (2012) Exploiting the mutanome for tumor vaccination. Cancer Res 72:1081–1091. doi:10.1158/0008-5472.CAN-11-3722
Abelin JG, Keskin DB, Sarkizova S, et al. (2017) Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 46:315–326. doi:10.1016/j.immuni.2017.02.007
Lundegaard C, Lamberth K, Harndahl M et al (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36:W509–W512. doi:10.1093/nar/gkn202
van Rooij N, van Buuren MM, Philips D et al (2013) Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 31:e439–e442. doi:10.1200/JCO.2012.47.7521
Duan F, Duitama J, Al Seesi S et al (2014) Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med 211:2231–2248. doi:10.1084/jem.20141308
Robbins PF, Lu Y-C, El-Gamil M et al (2013) Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 19:747–752. doi:10.1038/nm.3161
Kreiter S, Vormehr M, van de Roemer N et al (2015) Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520:692–696. doi:10.1038/nature14426
Snyder A, Makarov V, Merghoub T et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199. doi:10.1056/NEJMoa1406498
Bresciani A, Paul S, Schommer N et al (2016) T-cell recognition is shaped by epitope sequence conservation in the host proteome and microbiome. Immunology 148:34–39. doi:10.1111/imm.12585
Zou W (2006) Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol 6:295–307. doi:10.1038/nri1806
Reinherz EL (2015) αβ TCR-mediated recognition: relevance to tumor-antigen discovery and cancer immunotherapy. Cancer Immunol Res 3:305–312. doi:10.1158/2326-6066.CIR-15-0042
Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P (2013) Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun Arch 13:15
Novellino L, Castelli C, Parmiani G (2005) A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother 54:187–207. doi:10.1007/s00262-004-0560-6
Rammensee H, Bachmann J, Emmerich NP et al (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213–219
Vita R, Overton J a, Greenbaum J a et al (2015) The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43:D405–D412. doi:10.1093/nar/gku938
Seymour E, Damle R, Sette A, Peters B (2011) Cost sensitive hierarchical document classification to triage PubMed abstracts for manual curation. BMC Bioinform 12:482. doi:10.1186/1471-2105-12-482
Olsen L, Johan Kudahl U, Winther O, Brusic V (2013) Literature classification for semi-automated updating of biological knowledgebases. BMC Genom 14(Suppl 5):S14. doi:10.1186/1471-2164-14-S5-S14
Van den Eynde BJ, van der Bruggen P (1997) T cell defined tumor antigens. Curr Opin Immunol 9:684–693
Safran M, Dalah I, Alexander J, et al. (2010) GeneCards Version 3: the human gene integrator. Database 2010:baq020. doi:10.1093/database/baq020 (Oxford)
UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi:10.1093/nar/gku989
Forbes SA, Beare D, Gunasekaran P et al (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43:D805–D811. doi:10.1093/nar/gku1075
Olsen LR, Campos B, Barnkob MS et al (2014) Bioinformatics for cancer immunotherapy target discovery. Cancer Immunol Immunother 63:1235–1249. doi:10.1007/s00262-014-1627-7
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1006/jmbi.1990.9999
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/molbev/mst010
Nielsen M, Lundegaard C, Blicher T et al (2007) NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2:e796. doi:10.1371/journal.pone.0000796
Nielsen M, Justesen S, Lund O et al (2010) NetMHCIIpan-2.0—improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res 6:9. doi:10.1186/1745-7580-6-9
Zhang GL, Ansari HR, Bradley P et al (2011) Machine learning competition in immunology—prediction of HLA class I binding peptides. J Immunol Methods 374:1–4. doi:10.1016/j.jim.2011.09.010
Trolle T, Metushi IG, Greenbaum JA et al (2015) Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 31:2174–2181. doi:10.1093/bioinformatics/btv123
Zhang GL, Sun J, Chitkushev L, Brusic V (2014) Big data analytics in immunology: a knowledge-based approach. Biomed Res Int 2014:437987. doi:10.1155/2014/437987
Acknowledgements
This work was supported by The Danish Council for Independent Research Grant 4184-00211B (Lars Rønn Olsen), NIH Grant UO1 AI090043 and SU2C-AACR-DT13-14 (Ellis Reinherz) and Dana-Farber Cancer Institute, Cancer Vaccine Center Funds (Guang Lan Zhang and Vladimir Brusic).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Lars Rønn Olsen and Songsak Tongchusak contributed equally to the work.
Rights and permissions
About this article
Cite this article
Olsen, L.R., Tongchusak, S., Lin, H. et al. TANTIGEN: a comprehensive database of tumor T cell antigens. Cancer Immunol Immunother 66, 731–735 (2017). https://doi.org/10.1007/s00262-017-1978-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-017-1978-y